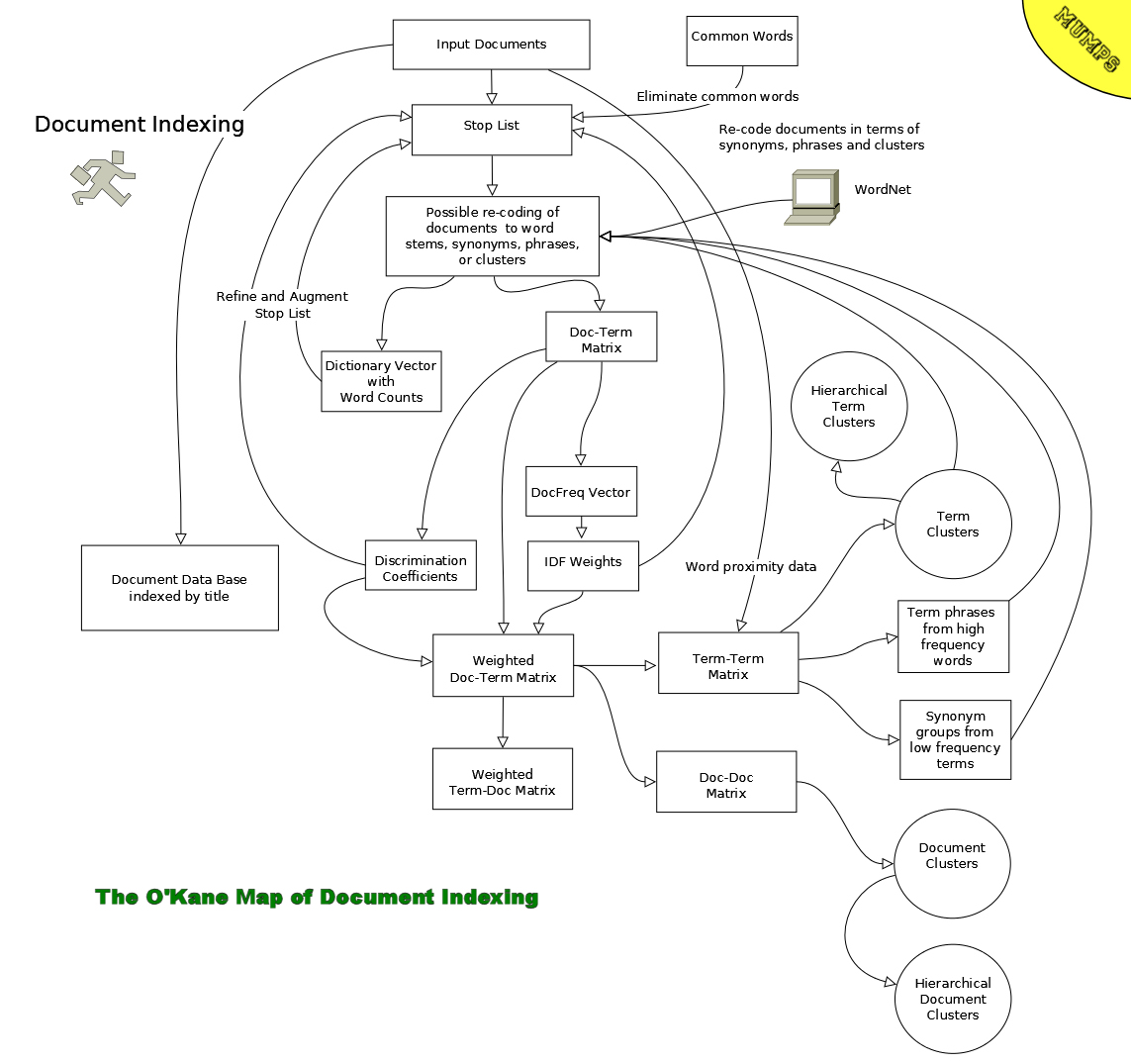
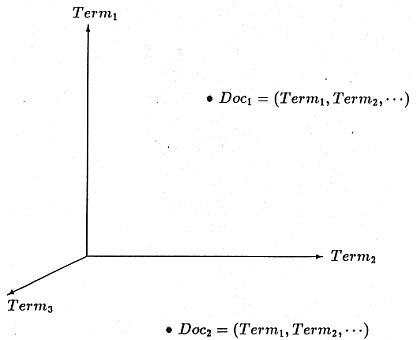
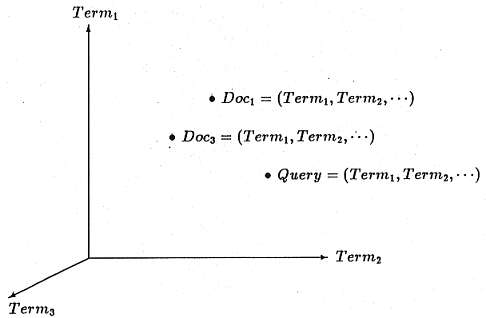
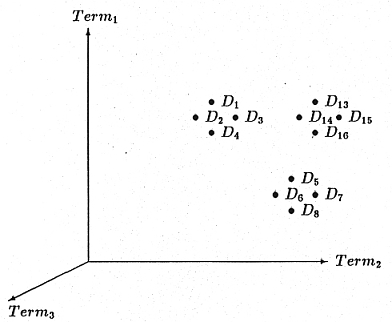
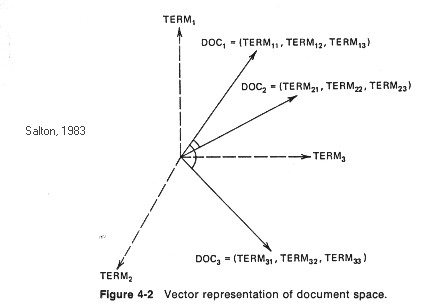
One popular approach to automatic document indexing,
the vector space model, views computer
generated document vectors as describing a hyperspace in which the number of dimensions
(axes) is equal to the number of indexing terms. This approach was originally proposed by
G. Salton:
Each document vector is a point in that space defined by the distance along the axis associated with each
document term proportional to the term's importance or significance in the document being represented.
Queries are also portrayed as vectors that define points in the document hyperspace. Documents whose points in the
hyperspace lie within an adjustable envelope of distance from the query vector point are retrieved.
The information storage and retrieval process involves converting user typed queries to query vectors and correlating these with
document vectors in order to select and rank documents for presentation to the user.
Most (IR) systems have been implemented in C, Pascal and C++, although these languages
provide little native support for the hyperspace model. Similarly, popular off-the-shelf legacy relational
data base systems are inadequate to efficiently represent or manipulate sparse document vectors in the
manner needed to effectively implement IR systems.
Document are viewed as points in a hyperspace whose
axes are the terms used in the document vectors.
The location of a document in the space is determined by
the degree to which the terms are present in a document.
Some terms occur several times while other occur not at all.
Terms also have weights associated with their content
indicating strength and this is factored into the equation
as well.
Query vectors are also treated as points in the hyperspace
and the documents that lie within a set distance of the query
are determined to satisfy the query.
Clustering involves identifying groupings of documents
and constructing a cluster centroid vector to
speed information storage and retrieval. Hierarchies of clusters
can also be constructed.
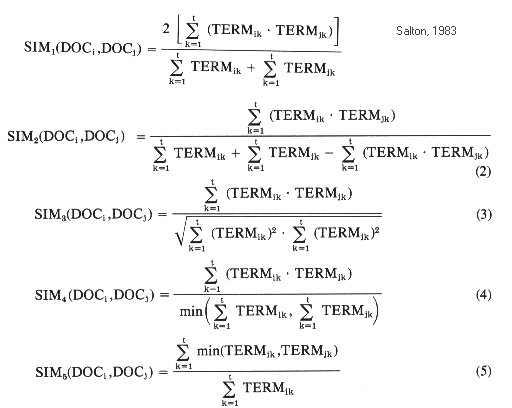
In the above, taken from Salton 1983, the Cosine is formula 3.
The formulae calculate the similarity between Doci and Docj
by examining the relationships between termi,k and termj,k
where termi,k is the weight of term k in document i and
termj,k is the weight of term k in document j.
Sim1 is known as the Dice coefficient and
Sim2 is known as the Jaccard coefficient
(see: Jaccard 1912, "The distribution of the flora of the alpine zone", New Phytologist 11:37-50).
Related Similarity Functions
See
Sam's String Metrics for a discussion of:
- Hamming distance
- Levenshtein distance
- Needleman-Wunch distance or Sellers Algorithm
- Smith-Waterman distance
- Gotoh Distance or Smith-Waterman-Gotoh distance
- Block distance or L1 distance or City block distance
- Monge Elkan distance
- Jaro distance metric
- Jaro Winkler
- SoundEx distance metric
- Matching Coefficient
- Dice.s Coefficient
- Jaccard Similarity or Jaccard Coefficient or Tanimoto coefficient
- Overlap Coefficient
- Euclidean distance or L2 distance
- Cosine similarity
- Variational distance
- Hellinger distance or Bhattacharyya distance
- Information Radius (Jensen-Shannon divergence)
- Harmonic Mean
- Skew divergence
- Confusion Probability
- Tau
- Fellegi and Sunters (SFS) metric
- TFIDF or TF/IDF
- FastA
- BlastP
- Maximal matches
- q-gram
- Ukkonen Algorithms
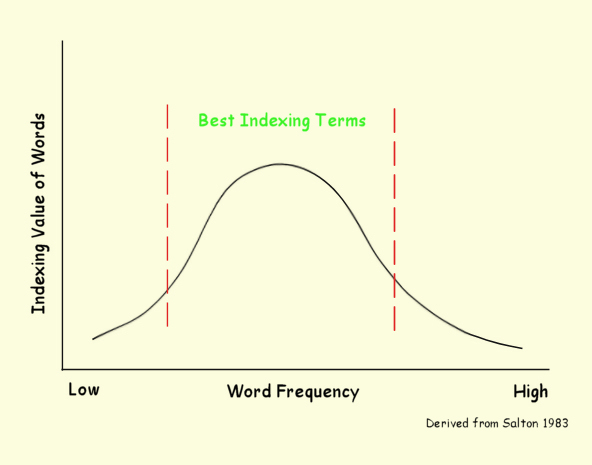
Assigning Word Weights
Words used for indexing vary in their ability to indicate content
and, thus, their importance as indexing terms.
Some words, such as "the", "and", "was" and so forth
are worthless as content indications and we eliminate them from consideration
immediately. Other words occur so infrequently that
they are also unlikely to be useful as indexing terms.
Other words, however, with middle frequency of occurrence
are candidates as indexing terms.
However, not all words a equally good index terms.
For example, the word "computer" in a collection of computer
science articles conveys very little information useful to
indexing the document since so many, if not all, the documents
contain the word. The goal here is to determine a metric
of the ability of a word to convey information.
In the following example, several weighting schemes are compared.
In the example, ^doc(i,w) is the number of times
term w occurs in document i; ^dict(w) is the number of
times term w occurs in the collection as a whole; ^df(w)
is the number of documents term w occurs in; NbrDocs
is the total number of documents in the collection; and the function $zlog()
is the natural logarithm. The operation "\" is integer division.
|
Normalize [normal.mps] Sun Dec 15 13:08:59 2002
1000 documents; 29942 word instances, 563 distinct words
^doc(i,w) Number times word w used in document i
^dict(w) Number times word w used in total collection
^df(w) Number of documents word w appears in
Wgt1 ^doc(i,w)/(^dict(w)/^df(w))
Wgt2 ^doc(i,w)*$zlog(NbrDocs/^df(w))+1
Wgt3 Wgt1*Wgt2+0.5\1
Word ^doc(i,w) ^dict(w) ^df(w) Wgt1 Wgt2 Wgt3 MCA
[1] Death of a cult. (Apple Computer needs to alter its strategy) (column)
apple 4 261 112 1.716 9.757 17 -1.1625
computer 4 706 358 2.028 5.109 10 -19.4405
mac 2 146 71 0.973 6.290 6 -0.0256
macintosh 4 210 107 2.038 9.940 20 -0.5855
strategy 2 79 67 1.696 6.406 11 -0.0592
[2] Next year in Xanadu. (Ted Nelson's hypertext implementations) Swaine, -
Michael.
document 3 114 68 1.789 9.065 16 0.0054
operate 3 269 184 2.052 6.078 12 -2.1852
[3] WordPerfect. (WordPerfect for the Macintosh 2.0) (evaluation) Taub, Er-
ic.
edit 2 111 77 1.387 6.128 8 -0.0961
frame 2 9 7 1.556 10.924 17 0.0131
import 2 29 19 1.310 8.927 12 0.0998
macintosh 3 210 107 1.529 7.705 12 -0.5855
macro 3 38 24 1.895 12.189 23 0.1075
outstand 1 10 9 0.900 5.711 5 0.0168
user 4 861 435 2.021 4.330 9 -26.8094
wordperfect 8 24 8 2.667 39.627 106 0.1747
[4] Radius Pivot for Built-In Video an Radius Color Pivot. (Hardware Revie-
w) (new Mac monitors)(includes related article on design of
built-in 3 35 29 2.486 11.621 29 0.0678
color 3 81 47 1.741 10.173 18 0.0809
mac 2 146 71 0.973 6.290 6 -0.0256
monitor 6 88 52 3.545 18.739 66 0.0946
resolution 2 50 32 1.280 7.884 10 0.0288
screen 2 92 62 1.348 6.561 9 0.0199
video 4 106 61 2.302 12.188 28 0.0187
[5] CrystalPrint Express. (Software Review) (high-speed desktop laser prin-
ter) (evaluation)
desk 2 127 76 1.197 6.154 7 -0.1062
engine 1 15 13 0.867 5.343 5 0.0282
font 4 111 37 1.333 14.187 19 0.6350
laser 3 61 27 1.328 11.836 16 0.2562
print 3 140 66 1.414 9.154 13 0.0509
[6] 4D Write, 4D Calc, 4D XREF. (Software Review) (add-ins for Acius' Four-
th Dimension database software) (evaluation)
add-in 2 97 38 0.784 7.540 6 0.5551
analysis 2 179 139 1.553 4.947 8 -0.8492
database 5 138 67 2.428 14.515 35 0.1832
midrange 1 7 6 0.857 6.116 5 0.0218
spreadsheet 2 75 44 1.173 7.247 9 0.1707
vary 1 7 6 0.857 6.116 5 0.0107
[7] ConvertIt! (Software Review) (utility for converting HyperCard stacks -
to IBM PC format) (evaluation)
converter 2 24 13 1.083 9.686 10 0.0698
doe 5 97 84 4.330 13.385 58 -0.1139
graphical 2 307 171 1.114 4.532 5 -2.4079
hypercard 4 25 13 2.080 18.371 38 0.1517
mac 2 146 71 0.973 6.290 6 -0.0256
map 2 17 10 1.176 10.210 12 0.1180
program 4 670 334 1.994 5.386 11 -15.4832
script 3 54 32 1.778 11.326 20 0.1239
software 3 913 449 1.475 3.402 5 -30.7596
stack 5 15 8 2.667 25.142 67 0.0700
[8] Reports 2.0. (Software Review) (Nine To Five Software Reports 2.0 repo-
rt generator for HyperCard 2.0) (evaluation)
hypercard 5 25 13 2.600 22.714 59 0.1517
print 3 140 66 1.414 9.154 13 0.0509
software 3 913 449 1.475 3.402 5 -30.7596
stack 2 15 8 1.067 10.657 11 0.0700
[9] Project-scheduling tools. (FastTrack Schedule, MacSchedule) (Software -
Review) (evaluation)
manage 2 318 174 1.094 4.497 5 -2.4884
[10] Digital Darkroom. (Software Review) (new version of image-processing s-
oftware) (evaluation)
apply 1 17 15 0.882 5.200 5 0.0317
digital 4 90 52 2.311 12.826 30 -0.0042
image 4 107 58 2.168 12.389 27 0.1422
palette 2 18 12 1.333 9.846 13 0.0660
portion 2 17 15 1.765 9.399 17 0.0295
software 4 913 449 1.967 4.203 8 -30.7596
text 2 55 46 1.673 7.158 12 0.0304
user 5 861 435 2.526 5.162 13 -26.8094
[11] CalenDAr. (Software Review) (Psyborn Systems Inc. CalenDAr desk access-
ory) (evaluation)
accessory 2 14 10 1.429 10.210 15 0.0540
desk 2 127 76 1.197 6.154 7 -0.1062
display 2 106 78 1.472 6.102 9 -0.1278
program 3 670 334 1.496 4.290 6 -15.4832
sound 2 14 8 1.143 10.657 12 0.1172
user 3 861 435 1.516 3.497 5 -26.8094
[12] DisplayServer II-DPD. (Hardware Review) (DisplayServer II video card f-
or using VGA monitor with Macintosh) (evaluation)
apple 4 261 112 1.716 9.757 17 -1.1625
card 2 99 56 1.131 6.765 8 0.0790
display 2 106 78 1.472 6.102 9 -0.1278
macintosh 3 210 107 1.529 7.705 12 -0.5855
monitor 6 88 52 3.545 18.739 66 0.0946
vga 2 91 62 1.363 6.561 9 0.0104
video 2 106 61 1.151 6.594 8 0.0187
[13] SnapJot. (Software Review) (evaluation) Gruberman, Ken.
capture 2 14 11 1.571 10.020 16 0.0271
image 3 107 58 1.626 9.542 16 0.1422
software 3 913 449 1.475 3.402 5 -30.7596
window 4 417 159 1.525 8.355 13 -3.4780
[14] Studio Vision. (Software Review) (Lehrman, Paul D.) (evaluation) Lehrm-
an, Paul D.
audio 1 8 6 0.750 6.116 5 0.0161
disk 3 234 121 1.551 7.336 11 -1.1468
edit 3 111 77 2.081 8.692 18 -0.0961
operate 2 269 184 1.368 4.386 6 -2.1852
portion 1 17 15 0.882 5.200 5 0.0295
requirement 2 87 76 1.747 6.154 11 -0.1203
sound 6 14 8 3.429 29.970 103 0.1172
user 3 861 435 1.516 3.497 5 -26.8094
[15] 70 things you need to know about System 7.0. (includes related article-
s on past reports about System 7.0, Adobe Type 1 fonts,
apple 3 261 112 1.287 7.568 10 -1.1625
communication 2 199 110 1.106 5.415 6 -0.6984
desk 2 127 76 1.197 6.154 7 -0.1062
disk 2 234 121 1.034 5.224 5 -1.1468
duplicate 1 10 9 0.900 5.711 5 0.0143
file 3 271 151 1.672 6.671 11 -1.3982
font 2 111 37 0.667 7.594 5 0.6350
memory 4 142 98 2.761 10.291 28 -0.2999
tip 1 8 6 0.750 6.116 5 0.0335
user 4 861 435 2.021 4.330 9 -26.8094
virtual 2 17 15 1.765 9.399 17 0.0424
[16] Data on the run. (Hardware Review) (palmtop organizers)(includes relat-
ed article describing the WristMac from Microseeds
character 2 25 17 1.360 9.149 12 0.0871
computer 4 706 358 2.028 5.109 10 -19.4405
data 3 415 226 1.634 5.462 9 -5.6011
database 2 138 67 0.971 6.406 6 0.1832
display 4 106 78 2.943 11.204 33 -0.1278
mac 3 146 71 1.459 8.935 13 -0.0256
ms_dos 2 98 65 1.327 6.467 9 0.0481
organize 1 19 17 0.895 5.075 5 0.0589
palmtop 1 6 5 0.833 6.298 5 0.0216
ram 2 145 93 1.283 5.750 7 -0.3992
review 2 265 238 1.796 3.871 7 -2.4234
rom 1 19 17 0.895 5.075 5 0.0374
software 4 913 449 1.967 4.203 8 -30.7596
transfer 2 66 44 1.333 7.247 10 0.0918
[17] High-speed, low-cost IIci cache cards. (includes related article on ca-
ching for other Mac models) (buyers guide)
cach 1 10 9 0.900 5.711 5 0.0127
cache 8 49 30 4.898 29.052 142 0.1613
card 6 99 56 3.394 18.294 62 0.0790
chip 2 117 67 1.145 6.406 7 -0.1153
high-speed 2 18 14 1.556 9.537 15 0.0352
memory 3 142 98 2.070 7.968 16 -0.2999
ram 2 145 93 1.283 5.750 7 -0.3992
[18] Mac, DOS and VAX file servers. (multiplatform file servers)(includes r-
elated articles on optimizing server
add-on 1 17 15 0.882 5.200 5 0.0374
apple 2 261 112 0.858 5.379 5 -1.1625
file 10 271 151 5.572 19.905 111 -1.3982
lan 2 98 51 1.041 6.952 7 0.0366
mac 4 146 71 1.945 11.580 23 -0.0256
macintosh 6 210 107 3.057 14.410 44 -0.5855
ms_dos 2 98 65 1.327 6.467 9 0.0481
netware 2 60 28 0.933 8.151 8 0.2314
network 6 571 222 2.333 10.030 23 -9.4287
ratio 1 18 16 0.889 5.135 5 0.0154
server 12 162 75 5.556 32.083 178 -0.1592
software 3 913 449 1.475 3.402 5 -30.7596
unix-based 1 15 13 0.867 5.343 5 0.0376
user 3 861 435 1.516 3.497 5 -26.8094
vax 2 28 14 1.000 9.537 10 0.1692
[19] Is it time for CD-ROM? (guide to 16 CD-ROM drives)(includes related ar-
ticles on using IBM-compatible CD-ROMs with the Mac,
audio 1 8 6 0.750 6.116 5 0.0161
cd-rom 9 31 13 3.774 40.085 151 0.1760
drive 9 249 129 4.663 19.431 91 -1.4872
macintosh 2 210 107 1.019 5.470 6 -0.5855
technology 2 335 220 1.313 4.028 5 -3.9304
[20] Silver platters that matter. (CD-ROM titles) (buyers guide)
availe 3 135 121 2.689 7.336 20 -0.4302
cd-rom 6 31 13 2.516 27.057 68 0.1760
hypercard 2 25 13 1.040 9.686 10 0.1517
library 2 44 30 1.364 8.013 11 0.1473
macintosh 2 210 107 1.019 5.470 6 -0.5855
|
In the example above, are document vectors for 20 documents (out of 1000) from
computer science trade publications of the mid-80's are shown. Several
weighting schemes are tried (see key at top). The MCA weight is the
Modified Centroid Algorithm calculation method to calculate the Term Discrimination weight (see below).
Inverse Document Frequency and Basic Vector Space
One of the simplest word weight schemes to implement is the
Inverse Document Frequency weight.
The IDF weight is the measure of how widely distributed
a term is in a collection. Low IDF weights mean that the term is
widely used while high weights indicate that the usage is more
concentrated. The IDF weight measures the weight of
a term in the collection as a whole, rather than the weight of
a term in a document. In individual document vectors,
the normalized frequency of occurrence of each term is multiplied by the
IDF to give a weight for the term in the particular document.
Thus, a term with a high frequency but a low IDF weight could
still be a highly weighted term in a particular document, and,
on the other hand, a term with a low frequency but a high
IDF weight could also be an important term in a given document.
The IDF weight for a term W in a collection of N documents is:
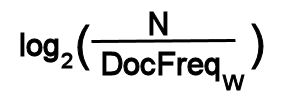
where DocFreqw is the number of documents in which term W
occurs.
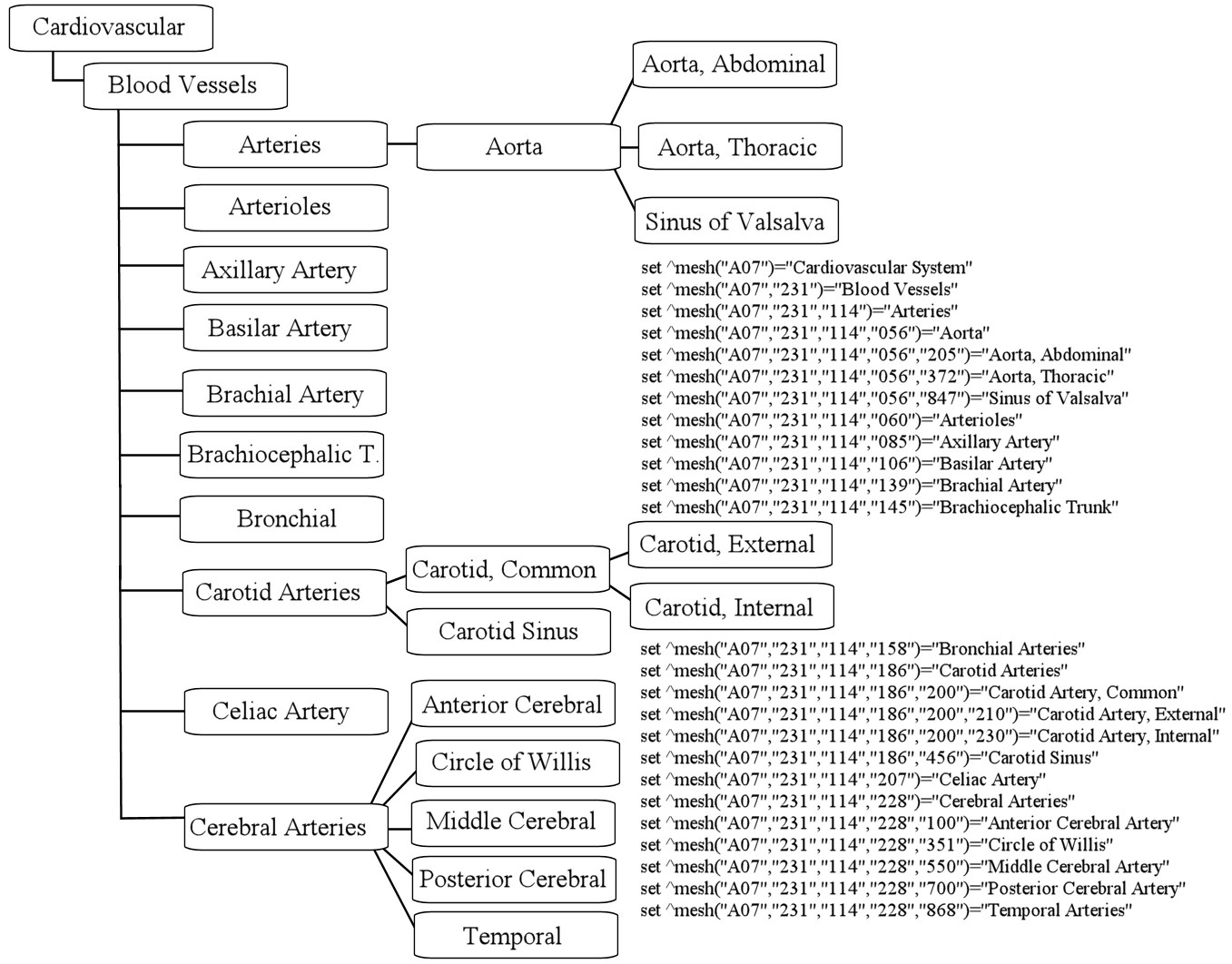
- OSU Medline Data Base IDF Weights
The IDF weights for the OSU Medline collection
were calculated after the words were processed by the stemming
function $zstem()
and the values are stored in the global array ^df(word)
for subsequent use and also printed to standard output.
The IDF weights for a recent run on the OSU
data base is here:
http://www.cs.uni.edu/~okane/source/ISR/medline.idf.sorted.gz.
Note: due to tuning parameters that set thresholds for the
construction of the stop list and other factors, different runs
on the data base will produce some variation in the
values displayed.
The weights range from lows such as:
0.189135 human
0.288966 and
0.300320 the
0.542811 with
0.737224 for
0.793466 was
0.867298 were
to highs such as:
12.590849 actinomycetoma
12.590849 actinomycetomata
12.590849 actinomycoma
12.590849 actinomyosine
12.590849 actinoplane
12.590849 actinopterygii
12.590849 actinoxanthin
12.590849 actisomide
12.590849 activ
12.590849 activationin
Note: for a given IDF value, the words are presented alphabetically.
The OSU Medline collection has many code words that appear only once.
- Wikipedia Data Base IDF Weights
Similarly, the Wikipedia
IDF weights were calculated
and the
results are in http://www.cs.uni.edu/~okane/source/ISR/wiki.idf.sorted.gz.
The weights range from lows such as:
1.61 further
1.62 either
1.63 especial
1.65 certain
1.65 having
1.67 almost
1.67 along
1.68 involve
1.68 receive
to highs such as:
9.87 altopia
9.87 alyque
9.87 amangkur
9.87 amarant
9.87 amaranthus
9.87 amarantine
9.87 amazonite
9.87 ambacht
9.87 ambiorix
Calculating IDF Weights
Calculating an IDF involves
first building a document-term
matrix (^doc(i,w)) where i is the document
number and w is a term. Each cell in the
document-term matrix will contain the count of
the number of times that the term occurs in the document).
Next, from the document-term matrix,
construct a document frequency vector (^df(w)) where
each element gives the number documents the term
w occurs in.
When the document frequency vector has been built,
the individual IDF values for each word can
be calculated.
The following assumes you are using the file
http://www.cs.uni.edu/~okane/source/ISR/medline.translated.txt.gz
which is a pre-processed version of the OSU Medline text.
The format of the file is:
- the code xxxxx115xxxxx followed by one blank
- the offset in the original text file of STAT- MEDLINE followed by a blank
- the document number followed by a blank
- one or more words, converted to lower case and stemmed by $zstem()
Example:
xxxxx115xxxxx 0 1 the bind acetaldehyde the active site ribonuclease alteration catalytic active ...
xxxxx115xxxxx 2401 2 reduction breath ethanol reading norm male volunteer follow mouth rins with ...
xxxxx115xxxxx 3479 3 does the blockade opioid receptor influence the development ethanol dependence ...
xxxxx115xxxxx 4510 4 drinkwatcher description subject and evaluation laboratory marker heavy drink ...
xxxxx115xxxxx 5745 5 platelet affinity for serotonin increased alcoholic and former alcoholic biological ...
xxxxx115xxxxx 7128 6 clonidine alcohol withdraw pilot study differential symptom respons follow ...
xxxxx115xxxxx 7915 7 bias survey drink habit this paper present data from genere populate survey ...
xxxxx115xxxxx 8653 8 factor associate with young adult alcohol abuse the present study examne ...
xxxxx115xxxxx 9862 9 alcohol and the elder relationship illness and smok group semi independent ...
xxxxx115xxxxx 11174 10 concern the prob our confidence statistic editorial
|
Calculating the IDF:
Step 1
- delete any previous instances of the ^doc() and
^df() global arrays.
Step 2
Loop:
- Read the next word into w from the file using the $zzScan
function.
- If $test is false, you have reached the end of
file and proceed to Step 3.
- If the word w is the beginning of
document token (xxxxx115xxxxx):
- read (use $zzScan) the offset and then the document number.
- Retain the document number as variable D.
- Store the offset at ^doc(D).
- Repeat the loop
- Check the word w against your stop list. If it
is in the stop list, Repeat the loop
- If ^doc(D,w) exists, increment it; if not,
create it with a value of 1.
- Repeat the loop
Step 3
- for each document i in ^doc(i)
- for each word w in ^doc(i,w)
- check if ^df(w) exists. If it does, increment
^df(w); if not create ^df(w) and store a value of one.
Step 4
- for each word in ^df(w)
- calculate its IDF using the value in ^df(w) (the number of documents the
word occurs in) and D, the total number of documents.
- store the results in a global array ^idf(w)
- write (re-direct stdout is easiest) to a file
the IDF value (2 decimal places are usually enough - see $justify),
a blank, followed by the word.
Step 5
- sort the file numerically according to IDF value (first field)
The results of the IDF procedure may be used to
enhance the stop list with words that have very low
values.
A basic program to create a document-term matrix and calculate IDF weights is:
|
#!/usr/bin/mumps
#+++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
#+
#+ Mumps ISR Software Library
#+ Copyright (C) 2006, 2008 by Kevin C. O'Kane
#+
#+ Kevin C. O'Kane
#+ okane@cs.uni.edu
#+
#+
#+ This program is free software; you can redistribute it and/or modify
#+ it under the terms of the GNU General Public License as published by
#+ the Free Software Foundation; either version 2 of the License, or
#+ (at your option) any later version.
#+
#+ This program is distributed in the hope that it will be useful,
#+ but WITHOUT ANY WARRANTY; without even the implied warranty of
#+ MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the
#+ GNU General Public License for more details.
#+
#+ You should have received a copy of the GNU General Public License
#+ along with this program; if not, write to the Free Software
#+ Foundation, Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
#+
#++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
# idf.mps February 22, 2008
kill ^df
kill ^dict
set %=$zStopInit("good") // loads stop list into a C++ container
open 1:"translated.txt,old"
if '$test write "translated not found",! halt
use 1
for do
. use 1
. set word=$zzScan
. if '$test break
. if word="xxxxx115xxxxx" set off=$zzScan,doc=$zzScan quit // new abstract
. if '$zStopLookup(word) quit // is "word" in the good list
. if $data(^doc(doc,word)) set ^doc(doc,word)=^doc(doc,word)+1
. else set ^doc(doc,word)=1
. if $data(^dict(word)) set ^dict(word)=^dict(word)+1
. else set ^dict(word)=1
use 5
close 1
set ^DocCount(1)=doc
for d="":$order(^doc(d)):"" do
. for w="":$order(^doc(d,w)):"" do
.. if $data(^df(w)) set ^df(w)=^df(w)+1
.. else set ^df(w)=1
for w="":$order(^df(w)):"" do
. set ^dfi(w)=$justify($zlog(doc/^df(w)),1,2)
. write $justify(^dfi(w),1,2)," ",w,!
write !
halt
|
The program also records the offset in the original file of the beginning of
the abstract and stores the IDF values in the vector ^dfi.
It also calculates a document frequency vector (number of
documents a term occurs in) ^df and a dictionary vector
(total frequency of occurrence for each word) ^dict.
Example results:
http://www.cs.uni.edu/~okane/source/ISR/medline.idf.sorted.gz
http://www.cs.uni.edu/~okane/source/ISR/medline.weighted-doc-vectors.gz
http://www.cs.uni.edu/~okane/source/ISR/medline.weighted-term-vectors.gz
http://www.cs.uni.edu/~okane/source/ISR/wiki.idf.sorted.gz
http://www.cs.uni.edu/~okane/source/ISR/wiki.weighted-doc-vectors.gz
http://www.cs.uni.edu/~okane/source/ISR/wiki.weighted-term-vectors.gz
Signal-noise ratio (see Salton83 links, pages 63-66)
Discrimination Coefficients (pages 66-71) and
Simple Automatic Indexing (pages Salton83, 71-75);
Willett 1985; Crouch 1988
The Term Discrimination factor measures the degree to which
a term differentiates one document from another. It is
calculated based on the effect a term has on overall
hyperspace density with and without a given term.
If the space density is greater when a term is removed
from consideration, that means the term was making documents
look less like one another (a good discriminator) while
terms whose remove decreases the density are poor
discriminators. The discrimination values for
a set of terms are similar to the values for the IDF weights but
not exactly.
The basic procedure calls for first calculating the average of pair-wise similarities
between all documents in the space.
Then for each word, the average of the pair-wise similarities of
all the documents is calculated without that word. The difference in the averages
is the term discrimination value for the word. When the average similarity
increases when a word is removed, the word was a good discriminator - it made
documents look less like one another. On the other hand,
if the average similarity decreased, the term was not a good
discriminator since it made the documents look more like one another.
In practice, this is an expensive weight to calculate unless
speed-up techniques are used.
The modified centroid algorithm (see Crouch 1987), is an attempt
to improve the speed of calculation. The exact
calculation, where all pairwise similarity values are calculated
each time, is of complexity of the order of
(N)(N-1)(w)(W) where N is the number of documents, W is the
number of words in the collection and w is
the average number of terms per document vector.
Crouch (1988) discusses several methods to speed this calculation.
The first of these, the approximate approach, consists of
calculating the similarities of the documents with a centroid
vector representing the collection as a whole rather
that pair-wise. This results in considerable simplification as the
number of similarities to be calculated drops from (N)(N-1)
to N.
Another modification, called the Modified Centroid Algorithm
is based on:
- Subtracting the original contributions to the sum of the
similarities of those documents containing some term W and replacing these
values with the similarities calculated between the
the centroid and the document vectors with W removed;
- Storing the original contributions to the total
similarity by each document in a vector for later use (rather than
recalculating this value); and
- Using an inverted list to identify those documents which
contain the indexing terms.
In the centroid approximation of discrimination coefficients,
a centroid vector is calculated. That is, a vector is created
whose individual components are the average usage of each
word in the vocabulary. A centroid vector is the average of
all the document vectors and, by analogy, is at the center of
the hyperspace.
When using a centroid vector,
rather than calculating all the pair-wise similarities
of each document with each other document,
the average similarity is calculated by comparing
each document with the centroid vector.
This improves the performance to a complexity on the order of (N)(w)(W).
As modified centroid algorithm (MCA) calculates the average
similarity, it stores the
contribution of each document to the total document density
(order n space required). When calculating the
effect of a term on the document space density, the MCA subtracts the
original contribution of those documents that contain the term
under consideration and re-adds the document's contribution
re-calculated without the contribution of the term under consideration.
Complexity is on the order of (DF)(w)(W) where
DF is the average number of documents in which a term occurs.
Finally, an inverted term-document matrix is used to
quickly identify those documents that contain terms of interest
rather than scanning through the entire document-term matrix looking
for documents containing a given term.
While the MCA method yields values that are only an approximation
of the exact method, the values are very similar in most cases and
the savings in time to calculate the coefficients is very significant.
Crouch (Crouch 1987) reports that the MCA method was on the
order of 527 times faster than the exact method on relatively small
data sets. Larger data sets yield even greater savings as the
time required for the exact method grows with the square of the
number of documents while the MCA method grows linearly with the
number of documents. The following is the basic MCA algorithm:
|
#!/usr/bin/mumps
#+++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
#+
#+ Mumps ISR Software Library
#+ Copyright (C) 2007, 2008 by Kevin C. O'Kane
#+
#+ Kevin C. O'Kane
#+ okane@cs.uni.edu
#+
#+
#+ This program is free software; you can redistribute it and/or modify
#+ it under the terms of the GNU General Public License as published by
#+ the Free Software Foundation; either version 2 of the License, or
#+ (at your option) any later version.
#+
#+ This program is distributed in the hope that it will be useful,
#+ but WITHOUT ANY WARRANTY; without even the implied warranty of
#+ MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the
#+ GNU General Public License for more details.
#+
#+ You should have received a copy of the GNU General Public License
#+ along with this program; if not, write to the Free Software
#+ Foundation, Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
#+
#++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
# discrim4.mps March 5, 2008
open 1:"discrim,new"
use 1
set D=^DocCount(1) // number of documents
kill ^mca
set t1=$zd1
#++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
# calculate centroid vector ^c() for entire collection and
# the sum of the squares (needed in cos calc but should only be done once)
#++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
for w="":$order(^dict(w)):"" do
. set ^c(w)=^dict(w)/D // centroid is composed of avg word usage
#++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
# Calculate total similarity of docs for all words (T) by
# calculating the sum of the similarities of each document with the centroid.
# Remember and store contribution of each document in ^dc(dn).
#++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
set T=0
for i="":$order(^doc(i)):"" do
. set cos=$zzCosine(^doc(i),^c)
. set ^dc(i)=cos // save contributions to total of each
. set T=cos+T // sum the cosines
#++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
# calculate similarity of doc space with words removed
#++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
for W="":$order(^dict(W)):"" do // for each word W
#++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
# For each document containing W, calculate sum of the contribution
# of the cosines of these documents to the total (T). ^dc(i) is
# the original contribution of doc i. Sum of contributions is stored in T1.
#++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
. set T1=0,T2=0
. for d="":$order(^index(W,d)):"" do //for each doc d containing W
.. set T1=^dc(d)+T1 // sum of orig contribution
.. kill ^tmp
.. for w1="":$order(^doc(d,w1)):"" do // make a copy of ^doc
... if w1=W quit // don't copy W
... set ^tmp(w1)=^doc(d,w1)
.. set T2=T2+$zzCosine(^tmp,^c) // sum of cosines without W
#++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
# subtract original contribution with W (T1) and add contribution
# without W (T2) and calculate r - the change, and store in ^mca(W)
#++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
# if old (T1) big and new (T2) small, density declines
. set r=T2-T1*10000\1
. write r," ",^dfi(W)," ",W,!
. set ^mca(W)=r
use 5
write $zd1-t1,!
close 1
halt
|
The following is a further refinement of the above. It
stores the sum of the squares of the components of the
centroid vector which are needed in the denominator of each
cosine calculation this eliminating this step
This version also eliminates the step where a copy
is made of the individual document vectors.
Overall, the changes noted above and implemented in the
program below can result in substantial time improvement.
On a test run on 10,000 abstracts from the Medline database, the
procedure above took 2,053 seconds while the one below took 378 seconds.
|
#!/usr/bin/mumps
#+++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
#+
#+ Mumps ISR Software Library
#+ Copyright (C) 2007, 2008 by Kevin C. O'Kane
#+
#+ Kevin C. O'Kane
#+ okane@cs.uni.edu
#+
#+
#+ This program is free software; you can redistribute it and/or modify
#+ it under the terms of the GNU General Public License as published by
#+ the Free Software Foundation; either version 2 of the License, or
#+ (at your option) any later version.
#+
#+ This program is distributed in the hope that it will be useful,
#+ but WITHOUT ANY WARRANTY; without even the implied warranty of
#+ MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the
#+ GNU General Public License for more details.
#+
#+ You should have received a copy of the GNU General Public License
#+ along with this program; if not, write to the Free Software
#+ Foundation, Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
#+
#++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
# discrim3.mps March 5, 2008
open 1:"discrim,new"
use 1
set D=^DocCount(1) // number of documents
set sq=0
kill ^mca
set t1=$zd1
#++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
# calculate centroid vector ^c() for entire collection and
# the sum of the squares (needed in cos calc but should only be done once)
#++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
for w="":$order(^dict(w)):"" do
. set ^c(w)=^dict(w)/D // centroid is composed of avg word usage
. set sq=^c(w)**2+sq // The sum of the squares is needed below.
#++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
# Calculate total similarity of doc for all words (T) space by
# calculating the sum of the similarities of each document with the centroid.
# Remember and store contribution of each document in ^dc(dn).
#++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
set T=0
for i="":$order(^doc(i)):"" do
. set x=0
. set y=0
. for w="":$order(^doc(i,w)):"" do
.. set d=^doc(i,w)
.. set x=d*^c(w)+x // numerator of cos(c,doc) calc
.. set y=d*d+y // part of denominator
#++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
# Calculate and store the cos(c,doc(i)).
# Remember in ^dc(i) the contribution that this document made to the total.
#++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
. if y=0 quit
. set ^dc(i)=x/$zroot(sq*y) // cos(c,doc(i))
. set T=^dc(i)+T // sum the cosines
#++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
# calculate similarity of doc space with words removed
#++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
for W="":$order(^dict(W)):"" do
#++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
# For each document containing W, calculate sum of the contribution
# of the cosines of these documents to the total (T). ^dc(i) is
# the original contribution of doc i. Sum of contributions is stored in T1.
#++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
. set T1=0,T2=0
. for i="":$order(^index(W,i)):"" do // row of doc nbrs for word
.. set T1=^dc(i)+T1 // use prevsly calc'd cos
#++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
# For each word in document i, recalculate cos(c,doc) but without word W
#++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
.. set x=0
.. set y=0
.. for w="":$order(^doc(i,w)):"" do
... if w'=W do // if W not w
.... set d=^doc(i,w)
.... set x=d*^c(w)+x // d*^c(w)+x
.... set y=d**2+y
.. if y=0 quit
.. set T2=x/$zr(sq*y)+T2 // T2 sums cosines without W
#++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
# subtract original contribution with W (T1) and add contribution
# without W (T2) and calculate r - the change, and store in ^mca(W)
#++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
# if old (T1) big and new (T2) small, density declines
. set r=T2-T1*10000\1
. write r," ",^dfi(W)," ",W,!
. set ^mca(W)=r
use 5
write "Time used: ",$zd1-t1,!
close 1
halt
|
Example results:
wiki.discrim.gz .
medline.discrim.sorted.gz .
Note: the discrimination coefficients output is in three columns:
the first is the coefficient times 10,000, the second is the IDF for the word
and the third is the word.
Basic Retrieval
Scanning the doc-term matrix
A simple program to scan the document-term matrix looking for documents
that have terms from a query vector. (Results based on 20,000 documents).
|
#!/usr/bin/mumps
# tq.mps Feb 27, 2008
kill ^query
write "Enter search terms: "
read a
if a="" halt
for i=1:1 do
. set b=$piece(a," ",i)
. if b="" break
. set b=$zn(b) // lower case, no punct
. set b=$zstem(b) // stem it
. set ^query(b)=""
if $order(^query(""))="" halt
for j="":$order(^query(j)):"" write j,!
set t1=$zd1
for i="":$order(^doc(i)):"" do
. set f=1
. for j="":$order(^query(j)):"" do
.. if '$d(^doc(i,j)) set f=0 break
. if f write i,?8,$extract(^t(i),1,70),!
write !,"Elapsed time: ",$zd1-t1,!
Enter search terms: epithelial fibrosis
epithelial
fibrosis
10001 Phosphorylation fails to activate chloride channels from cystic fibro
18197 Relationship between mammographic and histologic features of breast t
6944 Cyclic adenosine monophosphate-dependent kinase in cystic fibrosis tr
Elapsed time: 1
|
Scanning the term-doc matrix
A simple program to scan the term-document matrix looking for documents
that contain a search term.
|
#!/usr/bin/mumps
#+++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
#+
#+ Mumps ISR Software Library
#+ Copyright (C) 2008 by Kevin C. O'Kane
#+
#+ Kevin C. O'Kane
#+ okane@cs.uni.edu
#+
#+
#+ This program is free software; you can redistribute it and/or modify
#+ it under the terms of the GNU General Public License as published by
#+ the Free Software Foundation; either version 2 of the License, or
#+ (at your option) any later version.
#+
#+ This program is distributed in the hope that it will be useful,
#+ but WITHOUT ANY WARRANTY; without even the implied warranty of
#+ MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the
#+ GNU General Public License for more details.
#+
#+ You should have received a copy of the GNU General Public License
#+ along with this program; if not, write to the Free Software
#+ Foundation, Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
#+
#++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
# tqw.mps Feb 28, 2008
kill ^query
kill ^tmp
write "Enter search terms: "
read a
if a="" halt
for i=1:1 do
. set b=$piece(a," ",i)
. if b="" break
. set b=$zn(b) // lower case, no punct
. set b=$zstem(b) // stem it
. set ^query(b)=1
if $order(^query(""))="" halt
set q=0
for w="":$order(^query(w)):"" write w," " set q=q+1
write !
set t1=$zd1
for w="":$order(^query(w)):"" do
. for i="":$order(^index(w,i)):"" do
.. if $data(^tmp(i)) set ^tmp(i)=^tmp(i)+1
.. else set ^tmp(i)=1
for i="":$order(^tmp(i)):"" do
. if ^tmp(i)=q write i,?8,$j($zzCosine(^doc(i),^query),5,3)," ",$extract(^t(i),1,70),!
write !,"Elapsed time: ",$zd1-t1,!
Enter search terms: epithelial fibrosis
10001 0.180 Phosphorylation fails to activate chloride channels from cystic fibro
18197 0.291 Relationship between mammographic and histologic features of breast t
6944 0.323 Cyclic adenosine monophosphate-dependent kinase in cystic fibrosis tr
Elapsed time: 0
|
Weighted scanning the term-doc matrix
Similar to the above but all terms not required. Results sorted
by sum of weights of terms in the documents.
Note: the $job function returns the process id of the
running program. This is unique and it is used to
name a temporary file that contains the
unsorted results.
|
#!/usr/bin/mumps
# tqw1.mps Feb 27, 2008
kill ^query
kill ^tmp
write "Enter search terms: "
read a
if a="" halt
for i=1:1 do
. set b=$piece(a," ",i)
. if b="" break
. set b=$zn(b) // lower case, no punct
. set b=$zstem(b) // stem it
. set ^query(b)=""
if $order(^query(""))="" halt
set q=0
for w="":$order(^query(w)):"" write w," " set q=q+1
write !
set t1=$zd1
for w="":$order(^query(w)):"" do
. for i="":$order(^index(w,i)):"" do
.. if $data(^tmp(i)) set ^tmp(i)=^tmp(i)+^index(w,i)
.. else set ^tmp(i)=^index(w,i)
set fn=$job_",new"
open 1:fn // $job number is unique to this process
use 1
for i="":$order(^tmp(i)):"" do
. write ^tmp(i)," ",$extract(^t(i),1,70),!
close 1
use 5
set %=$zsystem("sort -n "_$job_"; rm "_$job)
write !,"Elapsed time: ",$zd1-t1,!
Enter search terms: epithelial fibrosis
epithelial fibrosis
9.02 Adaptation of the jejunal mucosa in the experimental blind loop syndr
9.02 Adherence of Staphylococcus aureus to squamous epithelium: role of fi
9.02 Anti-Fx1A induces association of Heymann nephritis antigens with micr
9.02 Anti-human tumor antibodies induced in mice and rabbits by "internal
9.02 Bacterial adherence: the attachment of group A streptococci to mucosa
9.02 Benign persistent asymptomatic proteinuria with incomplete foot proce
9.02 Binding of navy bean (Phaseolus vulgaris) lectin to the intestinal ce
9.02 Cellular and non-cellular compositions of crescents in human glomerul
9.02 Central nervous system metastases in epithelial ovarian carcinoma.
...
27.06 A new model system for studying androgen-induced growth and morphogen
27.06 Immunohistochemical observations on binding of monoclonal antibody to
27.7 Cyclic adenosine monophosphate-dependent kinase in cystic fibrosis tr
28.34 Relationship between mammographic and histologic features of breast t
28.98 Asbestos induced diffuse pleural fibrosis: pathology and mineralogy.
28.98 High dose continuous infusion of bleomycin in mice: a new model for d
28.98 Taurine improves the absorption of a fat meal in patients with cystic
33.81 Measurement of nasal potential difference in adult cystic fibrosis, Y
43.47 Are lymphocyte beta-adrenoceptors altered in patients with cystic fib
43.47 Lipid composition of milk from mothers with cystic fibrosis.
57.96 Pulmonary abnormalities in obligate heterozygotes for cystic fibrosis
Elapsed time: 0
|
Scripted test runs
Often it is better to break the indexing process into multiple steps.
The Mumps interpreter generally runs faster when the run-time symbol
table is not cluttered with many variable names.
Also, using a script can provide an easy way to set
parameters to the several steps from one central
code point.
Below is the bash script used to do the test
runs in this book. It invokes many individual Mumps
programs as well as other system resources such
as sort.
The script generally takes a considerable amount of time to
execute so it is often run under the control of nohup.
This permits the user to logoff and the script to continue
running. All output generated during execution that would
otherwise appear on your screed (stdout and stderr)
will instead be captured and written to the file nohup.out.
To invoke a script with nohup type:
nohup nice scriptName &
nohup will initiate the script and capture the output.
The nice command causes your script to run at a
slightly reduced priority thus giving interactive users
preference. The & causes the processes to run in the background
thus giving you a command prompt immediately (rather than
when the script is complete). Note: is you want to kill the
script,
type ps and then kill -9 pid where pid is
the process id of the script. You may also want to kill
the program currently running as killing the script
only stops the starting of additional tasks; tasks in execution
continue in execution.
Note that the Mumps interpreter always looks for QUERY_STRING
in the environment. Thus, if you create QUERY_STRING
and place in it parameters, Mumps will read these and create variables
with values as is the case when your program is invoked by
the web server:
QUERY_STRING="idf=$TT_MIN_IDF&cos=$TT_MIN_COS"
export QUERY_STRING
In the example above, query string is build and exported to the
environment. It contains two assignment clauses that
will result in the variables idf and cos
being created and initialized in Mumps before your program
begins execution. the bash variables TT_MIN_IDF
and TT_MIN_COS are established at the beginning of the script and
their values are substituted when QUERY_STRING is created.
Note the $'s - these cause the substitution and are required
by bash syntax.
|
#!/bin/bash
#+++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
#+
#+ Mumps Information Storage and Retrieval Software Library
#+ Copyright (C) 2006, 2008 by Kevin C. O'Kane
#+
#+ Kevin C. O'Kane
#+ okane@cs.uni.edu
#+
#+
#+ This program is free software; you can redistribute it and/or modify
#+ it under the terms of the GNU General Public License as published by
#+ the Free Software Foundation; either version 2 of the License, or
#+ (at your option) any later version.
#+
#+ This program is distributed in the hope that it will be useful,
#+ but WITHOUT ANY WARRANTY; without even the implied warranty of
#+ MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the
#+ GNU General Public License for more details.
#+
#+ You should have received a copy of the GNU General Public License
#+ along with this program; if not, write to the Free Software
#+ Foundation, Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
#+
#++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
# clear old nohup.out
cat /dev/null > nohup.out
# medline MedlineInterp.script March 9, 2008
TRUE=1
FALSE=0
# maximum number of documents to scan
MAX_DOCS=20000
# maximum word occurrence for stopselect.mps
MAX_WORDS=1000
# minumum word occurrence for stopselect.mps
MIN_WORDS=5
# minimum IDF value for doc-doc matrix
DD_MIN_IDF=7
# minimum DD cosine weight
DD_MIN_WGT=0.8
# minimum weight in weight.mps
MIN_WEIGHT=5
# minimum IDF in tt.mps
TT_MIN_IDF=5
# minimum IDF cosine
TT_MIN_COS=0.8
# minimum co-occurence tt count
TT_MIN_COUNT=10
# perform steps:
DO_ZIPF=$TRUE
DO_TT=$TRUE
DO_CONVERT=$TRUE
DO_DICTIONARY=$TRUE
DO_STOPSELECT=$TRUE
DO_IDF=$TRUE
DO_WEIGHT=$TRUE
DO_COHESION=$TRUE
DO_JACCARD=$TRUE
DO_TTCLUSTER=$TRUE
DO_DISCRIM=$TRUE
DO_DOCDOC=$TRUE
DO_CLUSTERS=$TRUE
DO_HIERARCHY=$TRUE
DO_TEST=$TRUE
if [ $DO_COHESION -eq $TRUE ]
then
DO_TT=$TRUE
fi
if [ $DO_JACCARD -eq $TRUE ]
then
DO_TT=$TRUE
fi
# delete any prior data bases
rm key.dat
rm data.dat
if [ $DO_CONVERT -eq $TRUE ]
then
echo "Convert data base format"
date
starttime.mps
QUERY_STRING="MAX=$MAX_DOCS"
export QUERY_STRING
echo "MAX documents to read $QUERY_STRING"
reformat.mps < osu.medline > rtrans.txt
stems.mps < rtrans.txt > translated.txt
echo "Conversion done - total time: `endtime.mps`"
echo
fi
if [ $DO_DICTIONARY -eq $TRUE ]
then
echo "Generate and sort word frequency list"
date
starttime.mps
dictionary.mps < translated.txt > dictionary.unsorted
sort -nr < dictionary.unsorted > dictionary.sorted
echo "Word frequency list done - total time: `endtime.mps`"
echo
fi
if [ $DO_ZIPF -eq $TRUE ]
then
echo "Zipf calculation"
date
starttime.mps
Zdictionary.mps < rtrans.txt | sort -nr | zipf.mps > medline.zipf
endtime.mps
echo
fi
echo "Count documents"
grep "xxxxx115" translated.txt | wc > DocStats
echo "Document grep result:"
cat DocStats
if [ $DO_STOPSELECT -eq $TRUE ]
then
echo "Select good words"
date
starttime.mps
QUERY_STRING="max=$MAX_WORDS&min=$MIN_WORDS"
export QUERY_STRING
echo "QUERY_STRING = $QUERY_STRING"
ls -l dictionary.sorted
stopselect.mps < dictionary.sorted > good
echo "Good word selection time: `endtime.mps`"
rm dictionary.unsorted
echo
fi
if [ $DO_IDF -eq $TRUE ]
then
echo "Generate and sort IDF weights"
date
starttime.mps
idf.mps > idf.unsorted
echo "idf done"
sort -n < idf.unsorted > idf.sorted
echo "IDF time: `endtime.mps`"
rm idf.unsorted
fi
if [ $DO_WEIGHT -eq $TRUE ]
then
echo "Create weighted doc vectors"
date
starttime.mps
# set minimum IDF*Freq value
QUERY_STRING="idfmin=$MIN_WEIGHT"
export QUERY_STRING
echo "QUERY_STRING = $QUERY_STRING"
weight.mps
echo "Weighting time: `endtime.mps`"
echo
fi
echo "Dump/restore"
echo "Old data base sizes:"
ls -lh key.dat data.dat
QUERY_STRING="file=weight.dmp"
export QUERY_STRING
starttime.mps
dump.mps
rm key.dat
rm data.dat
restore.mps
echo "New data base sizes:"
ls -lh key.dat data.dat
echo "Dump/restore time: `endtime.mps`"
echo
if [ $DO_TT -eq $TRUE ]
then
echo "Calculate and sort term-term matrix"
date
QUERY_STRING="min=$TT_MIN_COUNT&idf=$TT_MIN_IDF&cos=$TT_MIN_COS"
export QUERY_STRING
starttime.mps
tt.mps > tt
sort -n < tt > tt.sorted
echo "Term-term time: `endtime.mps`"
echo
if [ $DO_COHESION -eq $TRUE ]
then
echo "Calculate and sort cohesion matrix"
date
starttime.mps
cohesion.mps > cohesion
sort -nr < cohesion > cohesion.sorted
echo "Cohesion time: `endtime.mps`"
echo
fi
if [ $DO_JACCARD -eq $TRUE ]
then
echo "Calculate and sort jaccard term-term matrix"
date
starttime.mps
jaccard-tt.mps > jaccard-tt
sort -n < jaccard-tt > jaccard-tt.sorted
echo "Jaccard term time: `endtime.mps`"
echo
fi
if [ $DO_TTCLUSTER -eq $TRUE ]
then
echo "Calculate term clusters"
date
starttime.mps
clustertt.mps > cluster-tt
echo "Cluster term time: `endtime.mps`"
echo
fi
fi
if [ $DO_DISCRIM -eq $TRUE ]
then
echo "Calculate discrimination co-efficients"
date
starttime.mps
discrim3.mps
echo "discrim.mps time: `endtime.mps`"
sort -n < discrim > discrim.sorted
echo
fi
echo "Dump/restore"
date
echo "Old data base size:"
ls -lh key.dat data.dat
QUERY_STRING="file=discrim.dmp"
export QUERY_STRING
starttime.mps
dump.mps
rm key.dat
rm data.dat
restore.mps
echo "New data base size:"
ls -lh key.dat data.dat
echo "dump/restore end: `endtime.mps`"
echo
if [ $DO_DOCDOC -eq $TRUE ]
then
echo "Calculate document-document matrix"
date
starttime.mps
QUERY_STRING="wgt=$DD_MIN_WGT&min=$DD_MIN_IDF"
export QUERY_STRING
echo "QUERY_STRING = $QUERY_STRING"
docdoc5.mps > dd2
echo "Doc-doc time: `endtime.mps`"
echo
fi
if [ $DO_CLUSTERS -eq $TRUE ]
then
echo "Calculate clusters"
date
starttime.mps
cluster1.mps > clusters
echo "Cluster time: `endtime.mps`"
echo
fi
if [ $DO_HIERARCHY -eq $TRUE ]
then
echo "Calculate hierarchy"
date
starttime.mps
ttfolder.mps > ttfolder
echo "Hierarchy time: `endtime.mps`"
echo
echo "Calculate tables"
date
starttime.mps
tab.mps < ttfolder > tab
index.mps
echo "Hierarchy time: `endtime.mps`"
echo
fi
if [ $DO_TEST -eq $TRUE ]
then
echo "alcohol" > tstquery
echo "test query is alcohol"
medlineRetrieve.mps < tstquery
fi
|
Simple Retrieval
The following program reads in a set of query words into
a query vector and then calculates the cosines between
the query and the document vectors. It then prints out
the titles of the 10 documents with the highest cosine
correlations with the query.
Note: this program makes use of a synonym dictionary
which will be discussed below.
|
Basic Retrieval
|
|
#!/usr/bin/mumps
#+++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
#+
#+ Mumps ISR Software Library
#+ Copyright (C) 2005, 2007, 2008 by Kevin C. O'Kane
#+
#+ Kevin C. O'Kane
#+ okane@cs.uni.edu
#+
#+
#+ This program is free software; you can redistribute it and/or modify
#+ it under the terms of the GNU General Public License as published by
#+ the Free Software Foundation; either version 2 of the License, or
#+ (at your option) any later version.
#+
#+ This program is distributed in the hope that it will be useful,
#+ but WITHOUT ANY WARRANTY; without even the implied warranty of
#+ MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the
#+ GNU General Public License for more details.
#+
#+ You should have received a copy of the GNU General Public License
#+ along with this program; if not, write to the Free Software
#+ Foundation, Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
#+
#++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
# simpleRetrieval.mps Feb 28, 2008
open 1:"osu.medline,old"
if '$test write "osu.medline not found",! halt
write "Enter query: "
kill ^query
kill ^ans
for do // extract query words to query vector
. set w=$zzScanAlnum
. if '$test break
. set w=$zstem(w)
. set ^query(w)=1
write "Query is: "
for w="":$order(^query(w)):"" write w," "
write !
set time0=$zd1
for i="":$order(^doc(i)):"" do // calculate cosine between query and each doc
. if i="" break
. set c=$zzCosine(^doc(i),^query)
# If cosine is > zero, put it and the doc offset (^doc(i)) into an answer vector.
# Make the cosine a right justified string of length 5 with 3 digits to the
# right of the decimal point. This will force numeric ordering on the first key.
. if c>0 set ^ans($justify(c,5,3),^doc(i))=""
write "results:",!
set x=""
for %%=1:1:10 do
. set x=$order(^ans(x),-1) // cycle thru cosines in reverse (descending) order.
. if x="" break
. for i="":$order(^ans(x,i)):"" do
.. use 1 set %=$zseek(i) // move to correct spot in file primates.text
.. read a // skip STAT- MEDLINE
.. for k=1:1:30 do // the limit of 30 is to prevent run aways.
... use 1
... read a // find the title
... if $extract(a,1,3)="TI " use 5 write x," ",$extract(a,7,80),!
... if $extract(a,1,3)="AB " for do
.... use 5
.... write ?5,$extract(a,7,120),!
.... use 1
.... read a
.... if '$test break
.... if $extract(a,1,3)'=" " break
... if $extract(a,1,3)="STA" use 5 write ! break
write !,"Time used: ",$zd1-time0," seconds",!
which produces the following results on the first 20,000 abstracts:
Enter query: epithelial fibrosis
Query is: epithelial fibrosis
results:
0.393 Epithelial tumors of the ovary in women less than 40 years old.
From Jan 1, 1978 through Dec 31, 1983, 64 patients
with epithelial ovarian tumors, frankly malignant or
borderline, were managed at one institution. Nineteen
patients (29.7%) were under age 40. The youngest patient
was 19 years old. Nulliparity was present in 32% of
this group of patients. Of these young patients, 58%
had borderline epithelial tumors, compared to 13% of
patients over 40 years of age. Twenty-one percent of
the young patients were initially managed by unilateral
adnexal surgery. The overall cumulative actuarial survival
rate of all young patients was 93%. Young patients
with epithelial ovarian tumors tend to have earlier
grades of epithelial neoplasms, and survival is better
than that reported for older patients with similar
tumors.
0.367 Misdiagnosis of cystic fibrosis.
On reassessment of 179 children who had previously
been diagnosed as having cystic fibrosis seven (4%)
were found not to have the disease. The importance
of an accurate sweat test is emphasised as is the necessity
to prove malabsorption or pancreatic abnormality to
support the diagnosis of cystic fibrosis.
0.367 Are lymphocyte beta-adrenoceptors altered in patients with cystic fibrosis
1. Beta-adrenergic responsiveness may be decreased
in cystic fibrosis. In order to determine whether this
reflects an alteration in the human lymphocyte beta-receptor
complex, we studied 12 subjects with cystic fibrosis
(six were stable and ambulatory and six were decompensated,
hospitalized) as compared with 12 normal controls.
2. Lymphocyte beta-receptor mediated adenylate cyclase
activity (EC 4.6.1.1) was not decreased in the ambulatory
cystic fibrosis patients as compared with controls.
In contrast, decompensated hospitalized cystic fibrosis
patients demonstrated a significant reduction in beta-receptor
mediated lymphocyte adenylate cyclase activity expressed
as the relative increase over basal levels stimulated
by the beta-agonist isoprenaline compared with both
normal controls and stable ambulatory cystic fibrosis
patients (control 58 +/- 4%; ambulatory cystic fibrosis
patients 51 +/- 7%; decompensated hospitalized cystic
fibrosis patients 28 +/- 5%; P less than 0.05). 3.
Our data suggest that defects in lymphocyte beta-receptor
properties in cystic fibrosis patients may be better
correlated with clinical status than with presence
or absence of the disease state.
0.352 Measurement of nasal potential difference in adult cystic fibrosis, Young'
Previous work confirmed the abnormal potential difference
between the undersurface of the inferior nasal turbinate
and a reference electrode in cystic fibrosis, but the
technique is difficult and the results show overlap
between the cystic fibrosis and the control populations.
In the present study the potential difference from
the floor of the nose has therefore been assessed in
normal subjects, as well as in adult patients with
cystic fibrosis, bronchiectasis and Young's syndrome.
Voltages existing along the floor of the nasal cavity
were recorded. The mean potential difference was similar
in controls (-18 (SD 5) mv) and in patients with bronchiectasis
(-17 (6) mv) and Young's syndrome (-20 (6) mv). The
potential difference in cystic fibrosis (-45 (8) mv)
was significantly different from controls (p less than
0.002) and there was no overlap between the cystic
fibrosis values and values obtained in normal and diseased
controls. This simple technique therefore discriminates
well between patients with cystic fibrosis and other
populations, raising the possibility of its use to
assist in diagnosis.
0.342 Pulmonary abnormalities in obligate heterozygotes for cystic fibrosis.
Parents of children with cystic fibrosis have been
reported to have a high prevalence of increased airway
reactivity, but these studies were done in a select
young, healthy, symptomless population. In the present
study respiratory symptoms were examined in 315 unselected
parents of children with cystic fibrosis and 162 parents
of children with congenital heart disease (controls).
The cardinal symptom of airway reactivity, wheezing,
was somewhat more prevalent in cystic fibrosis parents
than in controls, but for most subgroups this increased
prevalence did not reach statistical significance.
Among those who had never smoked, 38% of obligate heterozygotes
for cystic fibrosis but only 25% of the controls reported
wheezing (p less than 0.05). The cystic fibrosis parents
who had never smoked but reported wheezing had lower
FEV1 and FEF25-75, expressed as a percentage of the
predicted value, than control parents; and an appreciable
portion of the variance in pulmonary function was contributed
by the interaction of heterozygosity for cystic fibrosis
with wheezing. For cystic fibrosis parents, but not
controls, the complaint of wheezing significantly contributed
to the prediction of pulmonary function (FEV1 and FEF25-75).
In addition, parents of children with cystic fibrosis
reported having lung disease before the age of 16 more
than twice as frequently as control parents. Other
respiratory complaints, including dyspnoea, cough,
bronchitis, and hay fever, were as common in controls
as in cystic fibrosis heterozygotes. These data are
consistent with the hypothesis that heterozygosity
for cystic fibrosis is associated with increased airway
reactivity and its symptoms, and that the cystic fibrosis
heterozygotes who manifest airway reactivity and its
symptoms may be at risk for poor pulmonary function.
0.323 Retroperitoneal fibrosis and nonmalignant ileal carcinoid.
The carcinoid syndrome and fibrosis are unusual but
identifiable disease processes. We report a rare case
of retroperitoneal fibrosis associated with an ileal
carcinoid in the absence of metastatic disease. The
literature is reviewed.
0.323 Cyclic adenosine monophosphate-dependent kinase in cystic fibrosis trachea
Cl-impermeability in cystic fibrosis (CF) tracheal
epithelium derives from a deficiency in the beta-adrenergic
regulation of apical membrane Cl- channels. To test
the possibility that cAMP-dependent kinase is the cause
of this deficiency, we assayed this kinase in soluble
fractions from cultured airway epithelial cells, including
CF human tracheal epithelial cells. Varying levels
of cAMP were used in these assays to derive both a
Vmax and apparent dissociation constant (Kd) for the
enzymes in soluble extracts. The cAMP-dependent protein
kinase from CF human tracheal epithelial cells has
essentially the same Vmax and apparent Kd as non-CF
human, bovine, and dog tracheal epithelial cells. Thus,
the total activity of the cAMP-dependent kinases and
their overall responsiveness to cAMP are unchanged
in CF.
0.313 Poor prognosis in patients with rheumatoid arthritis hospitalized for inte
Fifty-seven patients with rheumatoid arthritis (RA)
were treated in hospital for diffuse interstitial lung
fibrosis. Although interstitial fibrosis (either on
the basis of lung function tests or chest roentgenograms
or both) is fairly common among patients with RA, according
to this study interstitial fibrosis of sufficient extent
or severity to warrant hospitalization was rare: incidence
of hospitalization due to the lung disease in RA patients
was one case per 3,500 patient-years. Eight patients
had a largely reversible lung disease associated with
drug treatment (gold, D-penicillamine or nitrofurantoin.)
The remaining 49 had interstitial fibrosis of unknown
cause. Causes for hospitalization were respiratory
and general symptoms in 38, but infiltrations on routine
chest roentgenographic examinations alone in eleven
patients. Forty-five out of the 49 patients had crackles
on auscultation. The most typical findings in lung
function tests were restriction and a decreased diffusion
capacity. These 49 patients showed a poor prognosis,
with a median survival of 3.5 years and a five-year
survival rate of 39 percent.
0.291 Relationship between mammographic and histologic features of breast tissue
Mammograms and histologic slides of a group of 320
women who had breast symptoms and a biopsy without
cancer being found were reviewed. The mammographic
features assessed were the parenchymal pattern and
extent of nodular and homogeneous densities. In addition
to the pathologic diagnosis, the histologic features
assessed included epithelial hyperplasia and atypia,
intralobular fibrosis, and extralobular fibrosis. Among
premenopausal women, those with marked intralobular
fibrosis were more likely to have large (3+ mm) nodular
densities on the mammogram. Among postmenopausal women,
epithelial hyperplasia or atypia was related to having
nodular densities in at least 40% of the breast volume.
In both groups, marked extralobular fibrosis was related
to the presence of homogeneous density on the mammogram.
We conclude that mammographic nodular densities may
be an expression of lobular characteristics, whereas
homogeneous density may reflect extralobular connective
tissue changes.
0.290 Recent trends in the surgical treatment of endomyocardial fibrosis.
Several modifications of the traditional treatment
of endomyocardial fibrosis have been made based on
a personal experience of 51 surgical cases and on the
reports of others in the surgical literature during
the last decade. Description of these techniques and
the author's current concept of the pathological processes
are reported herein.
0.279 Vitamin A deficiency in treated cystic fibrosis: case report.
We describe a patient with cystic fibrosis and hepatic
involvement who, although on pancreatic extract, developed
vitamin A deficiency, night blindness, and a characteristic
fundus picture. All of these abnormalities were reversed
by oral vitamin A supplementation.
0.269 Epithelial ovarian tumor in a phenotypic male.
Laparotomy in a 41-year-old married man with non-treated
left cryptorchidism revealed female internal genitals
on the left side, and an epithelial ovarian tumor of
intermediate malignancy. Germinal malignancies are
frequent in intersexes, but non-germinal gonadal neoplasms
are rare. This is the second reported case of epithelial
ovarian tumor in intersexes, and the first case of
epithelial ovarian tumor in an intersex registered
as male.
Time used: 14 seconds
|
The above information storage and retrieval program is limited, however, because it sequentially
calculates the cosines between all documents and the query
vector. In fact, most documents contain no words in common with
the query vector and, consequently, their cosines are zero.
Thus, a possible speedup technique would be to only calculate the
cosines between the query and those documents that contain
at least one term in common with the query vector.
This
can be done by first constructing a vector of
document numbers of documents containing at least one
term in common with the query.
This is done in the following program as the query words are read
and processed. After processing a query word against the
stop list, synonym table, stemming and so on, if the resulting
term is in the vocabulary, add to a temporary vector ^tmp
those document numbers on the row from the term-document
matrix associated with the query term. When all query words
have been processed, the temporary vector
^tmp will contain, as indices, those document numbers
of documents that contain at least one query term.
While these documents represent, to some extent, a
response to the query, ranking the documents is important.
This could have been done somewhat simply by keeping
in ^tmp a count of the number of query terms
each document contained or it can now be calculated by
calculating a cosine or other suitable similarity
function between each of the document vectors whose
document numbers are in ^tmp and the query
vector ^query.
In the following, the cosine function is used to calculate the
similarity between the document vectors and the query vector.
As each cosine is calculated, it is stored along with the
value of ^doc(i) (where i^ans.
The purpose for doing this is to create a global array
ordered by cosine values as its first index and document identifiers
as its second index. That will allow the
results to be presented in descending cosine value order.
In order to avoid and ASCII sort of the numeric
cosine values, each cosine value is stored as an index
in a field of width 5 with three digits to the right of the decimal
point. This format insures that the first index will be in
numeric collating sequence order.
The second index of ^ans is the value of the
file offset pointer for the first line of the document
in the flat document file. Finally, the results are
presented in reverse cosine order (from high to low)
and the original documents at each cosine value are
printed (note: for a given cosine value, there may
be more than one document).
|
Faster Basic Retrieval
|
#!/usr/bin/mumps
#+++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
#+
#+ Mumps ISR Software Library
#+ Copyright (C) 2005, 2006, 2007, 2008 by Kevin C. O'Kane
#+
#+ Kevin C. O'Kane
#+ okane@cs.uni.edu
#+
#+
#+ This program is free software; you can redistribute it and/or modify
#+ it under the terms of the GNU General Public License as published by
#+ the Free Software Foundation; either version 2 of the License, or
#+ (at your option) any later version.
#+
#+ This program is distributed in the hope that it will be useful,
#+ but WITHOUT ANY WARRANTY; without even the implied warranty of
#+ MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the
#+ GNU General Public License for more details.
#+
#+ You should have received a copy of the GNU General Public License
#+ along with this program; if not, write to the Free Software
#+ Foundation, Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
#+
#++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
# fasterRetrieval.mps Feb 28, 2008
open 1:"osu.medline,old"
if '$test write "osu.medline not found",! halt
write "Enter query: "
kill ^query
kill ^ans
kill ^tmp
for do // extract query words to query vector
. set w=$zzScanAlnum
. if '$test break
. set w=$zstem(w)
. if '$data(^dict(w)) quit // skip unknown words
. set ^query(w)=1
write "Query is: "
for w="":$order(^query(w)):"" write w," "
write !
set time0=$zd1
# Find documents containing one or more query terms.
for w="":$order(^query(w)):"" do
. for d="":$order(^index(w,d)):"" set ^tmp(d)="" // retain doc id
for i="":$order(^tmp(i)):"" do // calculate cosine between query and each doc
. set c=$zzCosine(^doc(i),^query) // MDH cosine calculation
# If cosine is > zero, put it and the doc offset (^doc(i)) into an answer vector.
# Make the cosine a right justified string of length 5 with 3 digits to the
# right of the decimal point. This will force numeric ordering on the first key.
. if c>0 set ^ans($justify(c,5,3),^doc(i))=""
set x=""
for %%=1:1:10 do
. set x=$order(^ans(x),-1) // cycle thru cosines in reverse (descending) order.
. if x="" break
. for i="":$order(^ans(x,i)):"" do // get the doc offsets for each cosine value.
.. use 1 set %=$zseek(i) // move to correct spot in file primates.text
.. read a // skip STAT- MEDLINE
.. for k=1:1:30 do // the limit of 30 is to prevent run aways.
... use 1
... read a // find the title
... if $extract(a,1,3)="TI " use 5 write x," ",$extract(a,7,80),!
... if $extract(a,1,3)="AB " for do
.... use 5
.... write ?5,$extract(a,7,120),!
.... use 1
.... read a
.... if '$test break
.... if $extract(a,1,3)'=" " break
... if $extract(a,1,3)="STA" use 5 write ! break
write !,"Time used: ",$zd1-time0," seconds",!
yields the same results as above but takes less than 1 second.
|
Thesaurus construction (Salton83, pages 76-84)
It is possible to find connections between terms based on their
frequency of co-occurrence. Terms that co-occur frequently
together are likely to be related.
These relationships can indicate that the words
are synonyms or terms used to express a concept.
For example, a strong relationship between the words "artificial"
and "intelligence" in a computer science data base
is due to the phrase "artificial intelligence" which
names a branch of computing. In this case, the
relationship is not that of a synonym.
Similarly, in the medical data base
terms such as
"circadian" "rhythm" and
"vena" "cava" and
"herpes" "simplex" are concepts expressed as
more than one term.
On the other hand, as seen below, words like
"synergism" and "synergistic", "cyst" and
"cystic", "schizophrenia" and "schizophrenic",
"nasal" and "nose", and
"laryngeal" and "larynx"
are examples of synonym relationships.
In other cases, the relationship is not so tight
so as to be a full synonym but express a categorical
relationship such as "anesthetic" and "halothane",
"analgesia" and "morphine",
"nitrogen" and "urea", and
"nurse" and "personnel".
Regardless of the relationship, a thesaurus
table can be constructed giving a list of words that frequently co-occur.
With this information it is then possible to:
- augment queries with related words to improve recall;
- combine multiple related infrequently occurring terms into
broader, more frequently occurring categories terms; and,
- create middle frequency composite terms from otherwise unusable high
frequency component terms.
In its simplest form, we construct a square term-term
correlation matrix which gives the frequency of co-occurrence
of terms with one another. Thus, if some term A
occurs in 20 documents and if term B also occurs in these same
documents, the term-term correlation matrix cell for row A and column B
will have the entry 20. A term-term correlation matrix
lower diagonal matrix is the same as the upper diagonal
matrix since the relationship between term A and B is always the
same as the relationship between term B and A. The diagonal
itself is meaningless in that it is the correlation of a term with itself.
Calculating a complete term-term correlation matrix based on
all documents in a large collection can be very time
consuming.
In most cases, a term-term matrix
potentially contains many billions of elements
(the square of the number of vocabulary terms)
summed over the entire collection of documents.
In practice, however, it is only necessary to sample a representative
part of the total collection. That is, you can calculate a representative
matrix by looking at every fifth, tenth or twentieth
document, etc., depending on the total size of the collection.
As many collections can contain clusters of documents
concerning specific topics, it is generally better to sample
across all the documents than to only process each document in some leading
fraction of the collection.
Further, many words never occur with others, especially
in technical collections such as the
OSU medical data base. Thus, the term-term matrix
may be sparse.
More general topic collections, however,
will probably have matrices that are less sparse.
- Basic Term-Term Co-Occurrence Matrix
The basic term-term correlation matrix calculation
initially yields a sparse matrix of term-term counts.
The program proceeds by taking each document 'd' in the doc-term
matrix and selecting each term 'w' in document 'd'. For each
term 'w', it selects those terms 'w1' in 'd' which are alphabetically
greater than 'w'. For each pair of terms {w,w1}, it increments the
co-occurrence count
(or instantiates with a value of 1 if it did not exist) of the cell in
term-term matrix '^tt' at row 'w' and column 'w1'.
Effectively, this produces an upper diagonal
matrix but no values on the diagonal itself are calculated.
The term-term matrix is then examined and those elements
having a frequency of co-occurrence below
a threshold are deleted
(adjust this value depending on collection size.
The Cosine is calculated between the term vectors
and this becomes the value of the term-term matrix element.
The results sorted by cosine are here:
http://www.cs.uni.edu/~okane/source/ISR/medline.tt.sorted.gz.
The results sorted by frequency of co-occurrence
are here:
http://www.cs.uni.edu/~okane/source/ISR/medline.ttx.sorted.gz.
The complete term-term table is
http://www.cs.uni.edu/~okane/source/ISR/medline.ttw.gz
The sort commands for the cosine and co-occurrence sorts, respectively, are:
sort -n < tt > ttx.sorted
sort -n -k 2 < tt > ttx.sorted
This procedure can take several hours to execute on a large data base
even on fast machines.
|
#!/usr/bin/mumps
#+++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
#+
#+ Mumps ISR Software Library
#+ Copyright (C) 2005, 2006, 2008 by Kevin C. O'Kane
#+
#+ Kevin C. O'Kane
#+ okane@cs.uni.edu
#+
#+
#+ This program is free software; you can redistribute it and/or modify
#+ it under the terms of the GNU General Public License as published by
#+ the Free Software Foundation; either version 2 of the License, or
#+ (at your option) any later version.
#+
#+ This program is distributed in the hope that it will be useful,
#+ but WITHOUT ANY WARRANTY; without even the implied warranty of
#+ MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the
#+ GNU General Public License for more details.
#+
#+ You should have received a copy of the GNU General Public License
#+ along with this program; if not, write to the Free Software
#+ Foundation, Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
#+
#++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
# tt.mps March 8, 2009
kill ^tt
if '$data(cos) set cos=0.8
if '$data(idf) set idf=7
if '$data(min) set min=5
set t1=$zd1
for d="":$order(^doc(d)):"" do
. for w="":$order(^doc(d,w)):"" do
.. if ^dfi(w)<idf quit
.. for w1=w:$order(^doc(d,w1)):"" do
... if w1=w quit
... if ^dfi(w1)<idf quit
... if $data(^tt(w,w1)) set ^tt(w,w1)=^tt(w,w1)+1
... else set ^tt(w,w1)=1
for w1="":$order(^tt(w1)):"" do
. for w2="":$order(^tt(w1,w2)):"" do
.. if ^tt(w1,w2)<min kill ^tt(w1,w2) quit
.. set c=$zzCosine(^index(w1),^index(w2))
.. if c<cos kill ^tt(w1,w2) quit
.. set x=^tt(w1,w2),^tt(w1,w2)=c,^tt(w2,w1)=^tt(w1,w2)
.. write $justify(c,1,3)," ",$justify(x,6)," ",w1," ",w2,!
write "time=",$zd1-t1,!
|
The program to write the term-term matrix showing
for each word all related words:
|
#!/usr/bin/mumps
#+++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
#+
#+ Mumps ISR Software Library
#+ Copyright (C) 2005, 2006, 2008 by Kevin C. O'Kane
#+
#+ Kevin C. O'Kane
#+ okane@cs.uni.edu
#+
#+
#+ This program is free software; you can redistribute it and/or modify
#+ it under the terms of the GNU General Public License as published by
#+ the Free Software Foundation; either version 2 of the License, or
#+ (at your option) any later version.
#+
#+ This program is distributed in the hope that it will be useful,
#+ but WITHOUT ANY WARRANTY; without even the implied warranty of
#+ MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the
#+ GNU General Public License for more details.
#+
#+ You should have received a copy of the GNU General Public License
#+ along with this program; if not, write to the Free Software
#+ Foundation, Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
#+
#++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
# ttword.mps March 5, 2008
for w1="":$order(^tt(w1)):"" do
. write w1,?26
. set i=1
. for w2="":$order(^tt(w1,w2)):"" do
.. write w2," "
.. set i=i+1
.. if i#7=0 write !,?26
. if i#7'=0 write !!
|
Frequency

Word pair rank
In the above, the frequency of co-occurrence (number of times
two words co-occur together) for the whole OSU collection is plotted (no word pairs
eliminated).
The vertical axis represents the frequency scores and the horizontal axis
gives the rank of the term pair when sorted by frequency. That is,
the term pair with the highest score is the left most, the term pair with
the second highest frequency is second, and so on. As can be seen,
the frequency of co-occurrence drops off rapidly to a relatively constant,
slowly declining value. Thus, only a few term pairs in this
vocabulary, roughly the top 300, stand out as significantly more
likely to co-occur than the remainder of the possible
combinations.
The above calculation gives raw counts alone and does not take
into account the total number of times each term
occurs independently of the other. For example,
consider two terms that each occur with a high frequency.
It is more likely that these will co-occur together than
two terms whose individual frequencies of co-occurrence
are small.
Thus, the more sensitive Cosine based term-term correlation matrix can be calculated.
Unlike the
raw term-term co-occurrence count,
this method takes into account underlying
word usage frequency. The term-term co-occurrence
count from above, however, is biased to
show a greater co-occurrence for higher frequency
terms. Some lower frequency terms may have
usage profiles with one another that indicate
a greater similarity.
When the Cosines are graphed, the above has the following appearance: (right click then click "view image"
to see full size graph)
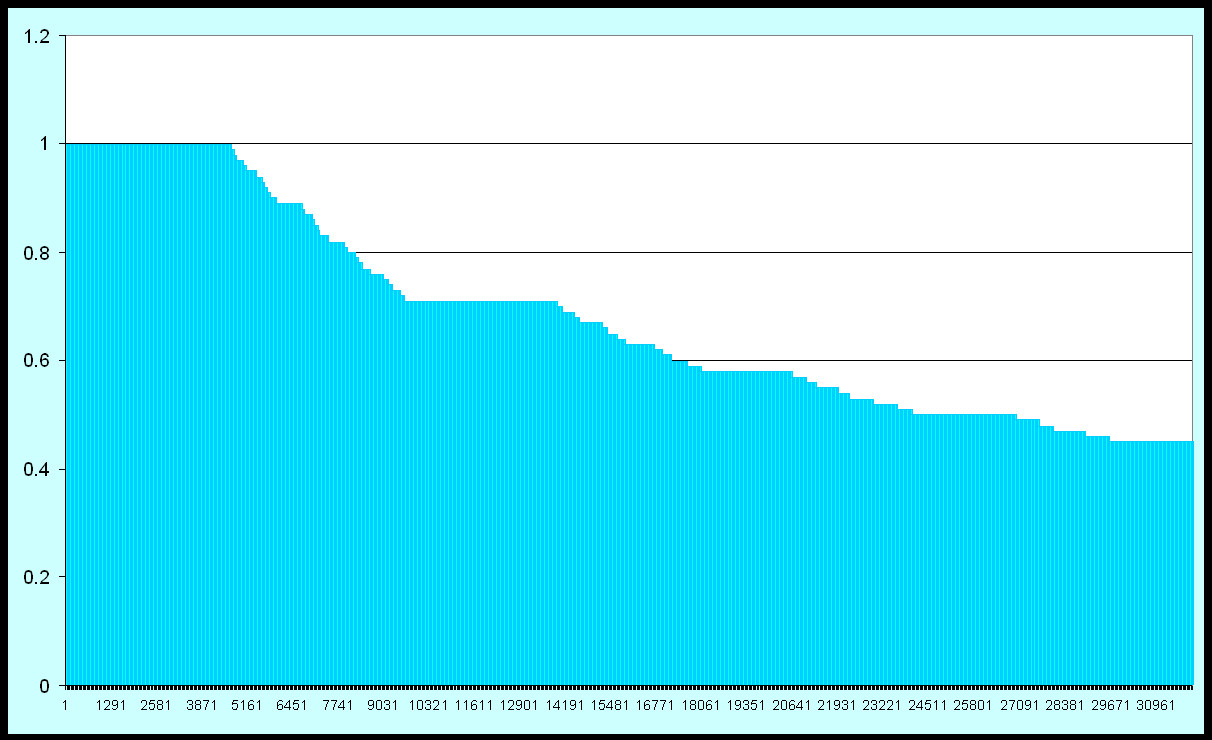
Again, the vertical axis is the cosine similarity between the term vectors
with a value of one indicating complete similarity and a value
of zero indicating no similarity between the vectors.
The difference in the graphs of the frequency based term-term
correlations and the Cosine based similarity calculation is due to the
larger number of significantly co-occurring low frequency terms which produce
a higher cosine value. These co-occurrences, due to the
lower underlying frequencies of the constituent terms,
ranked much lower in a frequency only calculation.
The several plateau's in the graph are due to the
rounding of the output to only 2 digits to the right of the
decimal point.
Alternatively, we can use the
the Jaccard similarity
(see above) formula which normalizes with respect to the
number of occurrences. The Jaccard values range between 0 and 1.
With this approach, if some term k occurs six times in the collection and some
term h occurs five times and they co-occur zero times,
the result is zero. If, however, they co-occur five times (the maximum),
and they each occur once in each per document, the result
is 5/(6+5-5) or 0.833. Similarly, if they each occur six times, once
per document and have a co-occurrence of six times, the result
is 6/(6+6-6) or 1.000. As can be seen, this formula, like the
cosine formula, takes into account the number of times each
term occurs independently of the number of co-occurrences.
The
Wikipedia results are here and the
OSU Medline results are here.
- Modified Basic Indexing - Position Specific
Another modification to improve term-term detection would
involve retaining during document scanning
the relative positions of the words with respect to one another
in the collection. Then, when calculating the term-term
matrix, proximity can be taken into account.
If you add a new matrix to the scanning code above
in addition to '^doc()' named
^p(DocNbr,Word,Position)
which in the third index position, retains for each
word in each document the position(s) which the word occurs
in relative to the beginning of the document (abstracts and
titles), it becomes possible to attenuate the
strength of co-occurrences by the proximity of the
co-occurrence.
Subsequently, the term-term calculation becomes: for each document
k, for each term i in k, for each other
term j where j is alphabetically higher
than i, for each position m
each position m of term i, and each position n
of term j, calculate and sum weights for ^tt(i,j) based on the
distance between the terms.
The distance is calculated with the formula:
set dd=$zlog(1/$zabs(m-n)*20+1)\1
which yields values of:
$zabs(m-n)=1 result=3
$zabs(m-n)=2 result=2
$zabs(m-n)=3 result=2
$zabs(m-n)=4 result=1
$zabs(m-n)=5 result=1
$zabs(m-n)=6 result=1
$zabs(m-n)=7 result=1
$zabs(m-n)=8 result=1
$zabs(m-n)=9 result=1
$zabs(m-n)=10 result=1
$zabs(m-n)=11 result=1
$zabs(m-n)=12 result=0
$zabs((m-n)=13 result=0
$zabs(m-n)=14 result=0
$zabs(m-n)=15 result=0
Thus, words immediately next to one another receive a score of three while words more than
eleven positions apart receive a score of zero.
For each pair {i,j} a third level index is summed which is the signed difference between m and n.
Eventually, a positive or negative value for this term indicates
a preference for which term appears first most often.
Values of zero or near zero indicate that the terms appear
in no specific order relative to one another.
The program then calculates a histogram giving the number of
term pairs at increasing scores (see link below).
For example, there were nearly 400,000 word combinations the sum
of whose scores was one.
Alternatively, there was one pair of words
whose co-occurrence score sum, calculated as above, was 1,732 (coron and artery).
The length of the histogram bars are based on the logarithm of the value
displayed to the left so as to make the graph more readable.
|
Proximity Weighted Term-Term Correlation Matrix
|
|
#!/usr/bin/mumps
#+++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
#+
#+ Mumps Information Storage and Retrieval Software Library
#+ Copyright (C) 2005 by Kevin C. O'Kane
#+
#+ Kevin C. O'Kane
#+ okane@cs.uni.edu
#+
#+
#+ This program is free software; you can redistribute it and/or modify
#+ it under the terms of the GNU General Public License as published by
#+ the Free Software Foundation; either version 2 of the License, or
#+ (at your option) any later version.
#+
#+ This program is distributed in the hope that it will be useful,
#+ but WITHOUT ANY WARRANTY; without even the implied warranty of
#+ MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the
#+ GNU General Public License for more details.
#+
#+ You should have received a copy of the GNU General Public License
#+ along with this program; if not, write to the Free Software
#+ Foundation, Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
#+
#++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
# proximity.mps March 13, 2008
set %=$zStopInit("good") // loads stop list into a C++ container
open 1:"translated.txt,old"
if '$test write "translated not found",! halt
use 1
set p=0
set doc=0
set M=20000
for do
. if doc>M set doc=doc-1 break
. use 1
. set word=$zzScan
. if '$test break
. if word="xxxxx115xxxxx" set off=$zzScan,doc=$zzScan,p=0 quit // new abstract
. if '$zStopLookup(word) quit // is "word" in the good list
. set p=p+1
. set ^p(doc,word,p)=""
use 5
close 1
set ^DocCount(1)=doc
# ttx term-term correlation matrix
# calculate term-term proximity coefficients within env words
kill ^tt //* delete any old term-term correlation matrix
write !!,"Term-Term Correlation [ttx.mps] ",$zd,!
#++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
# for each document k, sum the co-occurrences of words i and j
#++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
set Wgt=0
set Wgtx=0
Open 1:"ttx.tmp,new"
# for each document k
set k=""
for do
. set k=$order(^p(k))
. if k="" break
# for each term i in p k
. set i=""
. for do
.. set i=$order(^p(k,i))
.. if i="" break
# for each other term j in doc k
.. set j=i
.. for do
... set j=$order(^p(k,j))
... if j="" break
# for each position m of term i in doc k
... set m=""
... for do
.... set m=$order(^p(k,i,m))
.... if m="" break
# for each position n of term j in doc k
.... set n=""
.... for do
..... set n=$order(^p(k,j,n))
..... if n="" break
# calculate and store weight based on proximity
..... set dd=$zlog(1/$zabs(m-n)*20+1)\1
..... if dd<1 quit
..... if '$Data(^tt(i,j)) set ^tt(i,j)=dd,^tt(i,j,1)=n-m,Wgtx=Wgtx+1,Wgt=Wgt+dd
..... else set ^tt(i,j)=^tt(i,j)+dd,^tt(i,j,1)=^tt(i,j,1)+(n-m),Wgt=Wgt+dd
do graph
# normalize
set max=0
set i=""
for do
. set i=$order(^tt(i))
. if i="" break
. set j=i
. for do
.. set j=$order(^tt(i,j))
.. if j="" break
.. if ^tt(i,j)>max set max=^tt(i,j)
# set max=WgtFactor/100*dm\1
set max=max*.1\1
# build other diagonal matrix
set i=""
for do
. set i=$order(^tt(i))
. if i="" break
. set j=i
. for do
.. set j=$order(^tt(i,j))
.. if j="" break
.. if ^tt(i,j)<max kill ^tt(i,j)
.. else set ^tt(j,i)=^tt(i,j)
set i=""
set Wgt=Wgt/Wgtx
write !,"Average term-term correlation: ",Wgt,!
# write "Maximum term-term correlation: ",dm,!
# write "Weight factor percentage: ",WgtFactor,!
write "Discard term-term connections with weight less than: ",max,!
for do
. set i=$order(^tt(i))
. if i="" break
. if $order(^tt(i,""))="" quit
. write !!,"*** ",i,!," "
. set j=""
. for do
.. set j=$order(^tt(i,j))
.. if j="" break
.. if i=j quit
.. else do
... write:$x>65 !," "
... write " ",j,"[",^tt(i,j),"]"
... if j]i do
.... Use 1
.... write $Justify(^tt(i,j),6),?10,i," ",j,?50,^tt(i,j,1),!
.... Use 5
write !!
Close 1
set i=$zsystem("sort -n -r <ttx.tmp > ttx.rank") //* shell command
set i=$zsystem("rm ttx.tmp") //* shell command
write "Dump data: ",$zcd("ttx.dmp"),!
write $zd,!
halt
graph write !,"Graphs",!
kill ^hx
kill ^hxx
kill ^hr
set i=""
set k=0
for do
. set i=$order(^tt(i))
. if i="" break
. set j=""
. for do
.. set j=$order(^tt(i,j))
.. if j="" break
.. set k=k+1
.. set ^hr(k)=^tt(i,j)
set i=0
set dm=0
for do
. set i=$order(^hr(i))
. if i="" break
. if ^hr(i)>dm set dm=^hr(i)
for j=1:1:dm set ^hx(j)=0
set i=0
for do
. set i=$order(^hr(i))
. if i="" break
. set j=^hr(i)
. set ^hx(j)=^hx(j)+1
set hxmax=0
set j=$order(^hx(""))
for i=j:1:dm set ^hxx(i)=^hx(i) set ^hx(i)=$zlog(^hx(i)+1) if ^hx(i)>hxmax set hxmax=^hx(i)
write !,"Logarithmic Histogram Showing Number of Term Pairs for levels of Correlation Strength",!!
write " Corr Words",!!
set j=$order(^hx(""))
for i=j:1:dm do
. set k=^hx(i)/hxmax*100\1
. if ^hx(i)>0 do
.. set k=k+1
.. write $Justify(i,5),$j(^hxx(i),7)," "
.. for m=1:1:k write "*"
.. write !
quit
|
Full results are prox.gz
and
http://www.cs.uni.edu/~okane/source/ISR/medline.ttx.ranked
The first column is the sum of the proximity scores followed by the
words followed by the summation of the ordering factor. A negative number
indicates the words most often occur in reverse order to that shown.
As can be seen, a proximity based Term-Term matrix yields substantially
better results although it is considerably more expensive to
calculate in terms of time and space.
The example was calculated on the first 10,000 documents in the
data base.
- Term-Term clustering
Terms can be grouped into clusters using the results of the term-term
matrix above. In a single link clustering
system, a term is added to a cluster if it is related
to one term already in the cluster.
|
#!/usr/bin/mumps
#++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
#+
#+ Mumps Bioinformatics Software Library
#+ Copyright (C) 2004, 2005, 2006, 2008 by Kevin C. O'Kane
#+
#+ Kevin C. O'Kane
#+ anamfianna@earthlink.net
#+ okane@cs.uni.edu
#+
#+
#+ This program is free software; you can redistribute it and/or modify
#+ it under the terms of the GNU General Public License as published by
#+ the Free Software Foundation; either version 2 of the License, or
#+ (at your option) any later version.
#+
#+ This program is distributed in the hope that it will be useful,
#+ but WITHOUT ANY WARRANTY; without even the implied warranty of
#+ MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the
#+ GNU General Public License for more details.
#+
#+ You should have received a copy of the GNU General Public License
#+ along with this program; if not, write to the Free Software
#+ Foundation, Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
#+
#++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
# clustertt.mps March 8, 2009
kill ^clstr
kill ^x
open 1:"tmp,new"
use 1
for w1="":$order(^tt(w1)):"" do
. for w2=w1:$order(^tt(w1,w2)):"" do
.. write ^tt(w1,w2)," ",w1," ",w2,!
close 1
set %=$zsystem("sort -n -r < tmp > tmp.sorted")
open 1:"tmp.sorted,old"
set c=1
for do
. use 1
. read a // correlation word1 word2
. if '$test break
. set score=$p(a," ",1)
. set w1=$p(a," ",2)
. set w2=$p(a," ",3)
. if w1=w2 quit
. set f=1
# ^x() is a two dimensional array that contains, at the second level,
# a list of clusters to which the word (w1) belongs
# ^cluster() is the cluster matrix. Each row (s) is a cluster
# numbered 1,2,3 ... The second level is a list of the words
# in the cluster.
# The following
# code runs thru all the clusters first for w1 (w1) and
# adds w2 to those clusters w1 belongs to. It
# repeats the process for w2. If a word pair are not
# assigned to some cluster (f=1), they are assigned to a new
# cluster and the cluster number is incremented (c)
. if $d(^x(w1)) for s="":$order(^x(w1,s)):"" do
.. set ^clstr(s,w2)=""
.. set ^x(w2,s)=""
.. set f=0
. if $d(^x(w2)) for s="":$order(^x(w2,s)):"" do
.. set ^clstr(s,w1)=""
.. set ^x(w1,s)=""
.. set f=0
. if f do
.. set ^clstr(c,w1)="" set ^x(w1,c)=""
.. set ^clstr(c,w2)="" set ^x(w2,c)=""
.. set c=c+1
# print the clusters
close 1
use 5
write "number of clusters: ",c,!!
for cx="":$order(^clstr(cx)):"" do
. write cx," cluster",!
. for w1="":$order(^clstr(cx,w1)):"" do
.. use 5 write w1," "
. write !!
|
The results for a 20,000 abstract run are
http://www.cs.uni.edu/~okane/source/ISR/medline.cluster-tt
- Construction of Term Phrases (Salton83, pages 84-89)
Salton notes that recall can be improved if additional, related
terms are added to a query. Thus, a query for 'antenna' will
result in more hits in the data base if the related term
'aerial' is added. An increase in recall, however,
is often accompanied by a decrease in precision.
So, while 'aerial' is a commonly used synonym for
'antenna', as in 'television aerial', it can also refer to a dance move,
a martial arts move, skiing, various musical groups
and performances, and any activity that is done
at a height (e.g., aerial photography).
Thus, adding it to a query with antenna has the
potential to introduce many extraneous hits.
Identification of phrases, however, has the potential
to increase precision. These are composite terms of
high specificity - such as 'television aerial' noted
above. While both 'television' and 'aerial' individually
are broad terms, the phrase 'television aerial'
or 'television antenna' is quite specific.
When a phrase is identified, it becomes a single
term in the document vector.
Phrases can be identified by both syntactic and
statistical methods.
While techniques that take into account term
proximity
as well as co-occurrence such as suggested above,
Salton suggests the following simpler formula for construction of term phrases:
^Cohesion(i,j) = SIZE_FACTOR * (^tt(i,j)/(^dict(i)*^dict(j)))
The code to perform the cohesion calculation is here
|
#!/usr/bin/mumps
#+++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
#+
#+ Mumps ISR Software Library
#+ Copyright (C) 2006, 2008 by Kevin C. O'Kane
#+
#+ Kevin C. O'Kane
#+ anamfianna@earthlink.net
#+ okane@cs.uni.edu
#+
#+
#+ This program is free software; you can redistribute it and/or modify
#+ it under the terms of the GNU General Public License as published by
#+ the Free Software Foundation; either version 2 of the License, or
#+ (at your option) any later version.
#+
#+ This program is distributed in the hope that it will be useful,
#+ but WITHOUT ANY WARRANTY; without even the implied warranty of
#+ MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the
#+ GNU General Public License for more details.
#+
#+ You should have received a copy of the GNU General Public License
#+ along with this program; if not, write to the Free Software
#+ Foundation, Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
#+
#++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
# cohesion.mps March 25, 2010
# phrase construction
open 1:"tmp,new"
use 1
for i=$order(^tt(i)) do
. set j=i
. for do
.. set j=$o(^tt(i,j))
.. if j="" break
.. set c=^tt(i,j)/(^dict(i)*^dict(j))*100000\1
.. if c>0 write c," ",i," ",j,!
shell sort -nr < tmp > cohesion.results
shell rm tmp
|
The OSU Medline results are here:
http://www.cs.uni.edu/~okane/source/ISR/medline.cohesion.sorted.gz
and the Wikipedia results are here:
http://www.cs.uni.edu/~okane/source/ISR/wiki.cohesion.sorted.gz
.
Salton notes that this procedure can sometimes result in
unwanted connections such as 'Venetian blind' and
'blind Venetian'. For that reason, the aggregate relative
order of the terms, as shown above, can help to
decide when two terms are seriously linked.
That is, if
the order is strongly in favor of on term preceding
another, this indicates a probable phrase;
on the other hand, if the relative order is
in neither direction, this is probably not a
phrase.
Document-Document Matrices
It is also possible to construct Document-Document Matrices giving the
correlation between all documents which have significant similarities with
one another. Such a matrix can be used to generate document clusters and
for purposes of document browsing by permitting the user to
navigate related documents to the one being viewed.
That is, if a user finds on of the retrieved articles
of particular interest, a Document-Document matrix can be
used to quickly identify other documents related to the document of interest.
The source code is shown below
and the Wikipedia results are
http://www.cs.uni.edu/~okane/source/ISR/wiki.dd2.gz
and the OSU Medline results are
http://www.cs.uni.edu/~okane/source/ISR/medline.dd2.gz.
The program only calculates the cosines between
documents that share at least one term rather than
between all possible documents.
|
#!/usr/bin/mumps
#+++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
#+
#+ Mumps ISR Software Library
#+ Copyright (C) 2007, 2008 2010 by Kevin C. O'Kane
#+
#+ Kevin C. O'Kane
#+ anamfianna@earthlink.net
#+ okane@cs.uni.edu
#+
#+
#+ This program is free software; you can redistribute it and/or modify
#+ it under the terms of the GNU General Public License as published by
#+ the Free Software Foundation; either version 2 of the License, or
#+ (at your option) any later version.
#+
#+ This program is distributed in the hope that it will be useful,
#+ but WITHOUT ANY WARRANTY; without even the implied warranty of
#+ MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the
#+ GNU General Public License for more details.
#+
#+ You should have received a copy of the GNU General Public License
#+ along with this program; if not, write to the Free Software
#+ Foundation, Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
#+
#++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
#!/usr/bin/mumps
# docdoc5.mps March 27, 2010
write !!,"Document-document matrix ",$zd,!
if '$data(%1) set min=2
else set min=%1
if '$data(%2) set wgt=0.8
else set wgt=%1
kill ^dd
for w=$order(^idf(w)) do
. if ^idf(w)<min quit
. for d1=$order(^index(w,d1)) do
.. set d2=d1
.. for do
... set d2=$order(^index(w,d2))
... if d2="" break
... if $data(^dd(d1,d2)) quit
... set ^dd(d1,d2)=$zzCosine(^doc(d1),^doc(d2))
for d1=$order(^dd(d1)) do
. for d2=$order(^dd(d1,d2)) do
.. if ^dd(d1,d2)<wgt kill ^dd(d1,d2)
.. else set ^dd(d2,d1)=^dd(d1,d2)
for d1=$order(^dd(d1)) do
. write !,d1,": ",?10
. set k=0
. for d2=$order(^dd(d1,d2)) do
.. write d2,"(",$justify(^dd(d1,d2),1,2),") "
.. set k=k+1
.. if k#7=0 write !,?10
|
File and Document Clustering (Salton83, pages 215-222)
(see also:
advanced clustering)
-
Hierarchical Clustering
-
Web Document Clustering: A Feasibility Demonstration (pdf)
-
Document Clustering
-
Hierarchical Document Clustering (pdf)
-
A comparative study of generative methods for document clustering (pdf)
-
Demonstration of hierarchical document clustering (pdf)
-
Relationship-based Clustering and Cluster Ensembles for High-dimensional Data Mining,
Alexander Strehl
The program cluster.mps uses a single link clustering technique
similar to that used in the term clustering above.
The program generates and then reads a file of document-document
correlations sorted in reverse (highest to lowest)
correlation order.
|
#!/usr/bin/mumps
#++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
#+
#+ Mumps Bioinformatics Software Library
#+ Copyright (C) 2004, 2005, 2006, 2008 by Kevin C. O'Kane
#+
#+ Kevin C. O'Kane
#+ anamfianna@earthlink.net
#+ okane@cs.uni.edu
#+
#+
#+ This program is free software; you can redistribute it and/or modify
#+ it under the terms of the GNU General Public License as published by
#+ the Free Software Foundation; either version 2 of the License, or
#+ (at your option) any later version.
#+
#+ This program is distributed in the hope that it will be useful,
#+ but WITHOUT ANY WARRANTY; without even the implied warranty of
#+ MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the
#+ GNU General Public License for more details.
#+
#+ You should have received a copy of the GNU General Public License
#+ along with this program; if not, write to the Free Software
#+ Foundation, Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
#+
#++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
# cluster.mps March 9, 2008
kill ^clstr
kill ^x
open 1:"tmp,new"
use 1
for d1="":$order(^dd(d1)):"" do
. for d2=d1:$order(^dd(d1,d2)):"" do
.. write ^dd(d1,d2)," ",d1," ",d2,!
close 1
set %=$zsystem("sort -n -r < tmp > tmp.sorted")
open 1:"tmp.sorted,old"
set c=1
for do
. use 1
. read a // correlation doc1 doc2
. if '$test break
. set score=$p(a," ",1)
. set seq1=$p(a," ",2)
. set seq2=$p(a," ",3)
. if seq1=seq2 quit
. set f=1
# ^x() is a two dimensional array that contains, at the second level,
# a list of clusters to which the document number (seq1) belongs
# ^cluster() is the cluster matrix. Each row (s) is a cluster
# numbered 1,2,3 ... The second level is a list of the document
# numbers of those documents in the cluster. The following
# code runs thru all the clusters first for doc1 (seq1) and
# adds seq2 (doc2) to those clusters doc1 belongs to. It
# repeats the process for seq2 (doc2). If a doc pair are not
# assigned to some cluster (f=1), they are assigned to a new
# cluster and the cluster number is incremented (c)
. if $d(^x(seq1)) for s="":$order(^x(seq1,s)):"" do
.. set ^clstr(s,seq2)=""
.. set ^x(seq2,s)=""
.. set f=0
. if $d(^x(seq2)) for s="":$order(^x(seq2,s)):"" do
.. set ^clstr(s,seq1)=""
.. set ^x(seq1,s)=""
.. set f=0
. if f do
.. set ^clstr(c,seq1)="" set ^x(seq1,c)=""
.. set ^clstr(c,seq2)="" set ^x(seq2,c)=""
.. set c=c+1
# print the clusters
close 1
open 1:"osu.medline,old"
if '$test write "missing translated.txt",! halt
use 5
write "number of clusters: ",c,!!
for cx="":$order(^clstr(cx)):"" do
. use 5 write cx," cluster",!
. for seq1="":$order(^clstr(cx,seq1)):"" do
.. use 1 set %=$zseek(^doc(seq1)) read title
.. use 5 write $e(title,1,120),!
. use 5 write !
|
The results for OSU Medline are here:
http://www.cs.uni.edu/~okane/source/ISR/medline.clusters.gz
and the Wikipedia results are here:
http://www.cs.uni.edu/~okane/source/ISR/wiki.clusters.gz.
Web Page Access -
Building a Simple Keyword Based Logical Expression Server Page
The following will outline the process to build an interactive
web page to access the data.
At this point, we assume that the data base has been processed
and the appropriate global array vectors and matrices exist.
The following program will access the data base:
|
#!/usr/bin/mumps
#++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
#+
#+ Mumps ISR Software Library
#+ Copyright (C) 2000, 2005, 2006 by Kevin C. O'Kane
#+
#+ Kevin C. O'Kane
#+ anamfianna@earthlink.net
#+ okane@cs.uni.edu
#+
#+
#+ This program is free software; you can redistribute it and/or modify
#+ it under the terms of the GNU General Public License as published by
#+ the Free Software Foundation; either version 2 of the License, or
#+ (at your option) any later version.
#+
#+ This program is distributed in the hope that it will be useful,
#+ but WITHOUT ANY WARRANTY; without even the implied warranty of
#+ MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the
#+ GNU General Public License for more details.
#+
#+ You should have received a copy of the GNU General Public License
#+ along with this program; if not, write to the Free Software
#+ Foundation, Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
#+
#++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
# webFinder.mps March 23, 2008
html Content-type: text/html &!&!
set t=$zd1
html <html><body bgcolor=silver>
if '$data(query) set query=""
html <center><img src=http://sidhe.cs.uni.edu/moogle.gif border=0><br>
html <form name="f1" method="get" action="webFinder.cgi">
html <input type="text" name="query" size=50 maxlength=128 value="&~query~">
html &nbsp <input type="submit" value="Search">
html </form>
html <form name="f1" method="get" action="webFinder.cgi">
html <input type="hidden" name="query" value="###">
html &nbsp <input type="submit" value="I'm Feeling Sick">
html </form></center>
write !
if query="" write "</body></html>",! halt
if query="###" do
. set w=""
. for i=1:1 do
.. set w=$order(^dict(w))
.. if w="" break
. set j=$r(i-1)
. set w=""
. for i=1:1:j do
.. set w=$order(^dict(w))
. set query=w
kill ^d
kill ^query
do $zwi(query)
set wx=0
for w=$zwp do
. if w?.P continue
. set ^query(w)=1
. for d="":$order(^index(w,d)):"" set ^d(d)=""
Set i=$zwi(query)
set exp=""
for w=$zwp do
. if w="" break
. if $find("&()",w) set exp=exp_w continue
. if w="|" set exp=exp_"!" continue
. if w="~" set exp=exp_"'" continue
. set exp=exp_"$d(^doc(d,"""_w_"""))"
write "<br>",exp,"<br>",!
kill ^dx
set max=0
set count=0
for d="":$order(^d(d)):"" do
. set $noerr=1 // corrected for interpreter use Apr 22, 2009
. set %=@exp
. if $noerr<0 write "Query parse error.</body></html>",! halt
. if %>0 do
.. set C=$zzCosine(^query,^doc(d))
.. set ^dx(C,d)=""
.. set count=count+1
write count," pages found - top 10 shown<hr><tt><pre>",!
set d=""
set i=0
open 1:"translated.txt,old"
if '$test write "translated.txt not found",! halt
for do
. if i>10 break
. set d=$order(^dx(d),-1)
. if d="" break
. for dd="":$order(^dx(d,dd)):"" do
.. set i=i+1
.. if i>10 break
.. write $j(d,6,3)," "
.. write "<a href=display.cgi?ref=",dd,">"
.. use 1 do $zseek(^doc(dd)) read title use 5
.. write $j(dd,6)," ",$extract(title,1,90),"</a>",!
write "<pre><p>Time used: ",$zd1-t,"<br>",!
html </body></html>
kill ^d
kill ^dx
halt
|
In the program, the interpreter decodes the
incoming environment variable QUERY_STRING set
by the web server. It instantiates variable
names with values as found in "xxx=yyy" figures
contained in QUERY_STRING. In particular, this
application receives a variable named "query"
which is either empty, the value "###" or
an optionally parenthesized logical expression
involving keywords.
In the case where "query" is missing or empty, only the
form text box is returned to the browser. In the
case where the value of "query" is "###", a random word is selected
from the vocabulary and this work becomes the value of
"query"
The value of "query" is processed first to extract all the
words provided. A global array vector named "query" is
constructed with the words as indices. For each word,
all the document numbers associated with the word in
^index() are fetched and stored in a temporary
vector ^d(). When processing of all words is complete,
the vector ^query() contains the words and the
vector ^d() contains the document numbers of all
documents that have one or more words in common with
the query.
Next, the query is rescanned and a Mumps string is built.
For each word in the query, a entry of the form:
$d(^doc(d,"word"))
is constructed where the value of the word is enclosed in
quotes. These are connected to other similar
expressions by not (~), and (&), or (!), and parentheses. Note:
the input vertical bar character (|) is converted to the Mumps exclamation
point "or" character (!). For example, query:
(ducks & chickens) | (rabbits & foxes)
becomes:
($d(^doc(d,"ducks"))&$d(^doc(d,"chickens")))!($d(^doc(d,"rabbits"))&$d(^doc(d,"foxes")))
Note: parsing of Mumps expressions is always left to right (no
precedence) unless parentheses are used.
Once the query has been re-written, the program cycles through
each document number in ^d(). For each document number "d",
the expression built from the query is executed interpretively
and the result, zero or greater than zero, determines
if the document should be processed further.
If the result is greater than zero, the Cosine of the
document vector and the query is calculated and stored in
the temporary vector ^dx(cosine,docNbr). This two part
index is used so that the same Cosine value can have multiple document
numbers linked to it.
Finally, the document titles (^t()) are printed in reverse cosine order.
Each title is expressed as a link to the program display.cgi
which will ultimately display the abstract.
The program produces output such as
(OSU Medline example):
A live (or dead - depends on the server) version can
be seen
by clicking here .
Unfortunately, the program above depends upon the user to be
familiar with the vocabulary used in the data base.
A user who spells a word incorrectly or uses a similar
but different synonym is out of luck.
However, there are several ways to
improve data base navigation.
The program above can be extended with a front end that permits point-and-click browsing
for terms and term combinations.
In order to do this, we used the results of the term-term matrix above
with a lower threshold (thereby increasing the number of possible
word combinations).
The term-term matrix was used as input to a program that
produced an alphabetic two level hierarchical organization of the terms
that became input for a point-and-click web based
display procedure.
|
#!/usr/bin/mumps
#++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
#+
#+ Mumps ISR Software Library
#+ Copyright (C) 2000, 2005, 2008 by Kevin C. O'Kane
#+
#+ Kevin C. O'Kane
#+ anamfianna@earthlink.net
#+ okane@cs.uni.edu
#+
#+
#+ This program is free software; you can redistribute it and/or modify
#+ it under the terms of the GNU General Public License as published by
#+ the Free Software Foundation; either version 2 of the License, or
#+ (at your option) any later version.
#+
#+ This program is distributed in the hope that it will be useful,
#+ but WITHOUT ANY WARRANTY; without even the implied warranty of
#+ MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the
#+ GNU General Public License for more details.
#+
#+ You should have received a copy of the GNU General Public License
#+ along with this program; if not, write to the Free Software
#+ Foundation, Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
#+
#++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
# ttfolder.mps January 2, 2008
for i=0:1:25 do
. set a=$c(97+i)
. set w1=$order(^dfi(a))
. if $e(w1,1,1)'=a continue
. write a,!
. write """blank.html"" target=right",!!
. for w1=a:$order(^dfi(w1)):"" do
.. if $e(w1,1,1)'=a break
.. if ^dfi(w1)<3 quit
.. write ". ",w1,!
.. write ". ""blank.html"" target=right",!!
.. write ".. ",w1,!
.. write ".. ""webFinder.cgi?query=",$zh(w1),""" ",!!
.. if '$data(^tt(w1)) quit
.. for w2="":$order(^tt(w1,w2)):"" do
... write ".. ",w1," & ",w2,!
... write ".. ""webFinder.cgi?query=",$zh(w1_"&"_w2),""" ",!!
|
First, we use a
program (above) that extracts term pairs from the term-term matrix.
The OSU Medline results are here
http://www.cs.uni.edu/~okane/source/ISR/medline.ttfolder.gz.
The output from the above is input to a program that produces dynamic, web based
folders. Each entry is two lines with a blank line following.
The first line is the text to be displayed and the second
line is the action or page to be performed if the
text is clicked. The number of dots preceding each
line determines the level in the hierarchy of the entry.
The file is organized at the highest level by letters of the
alphabet (a through z) and the n by words beginning
with that letter. Beneath each word is a list of
words with a significant correlation to the higher
term, as determined by the term-term matrix.
The program that reads the output of the above and builds the hierarchy is here:
|
#!/usr/bin/mumps
# ++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
#
# Information Storage and Retrieval Indexer
# Copyright (C) 2000, 2005, 2006, 2008 by Kevin C. O'Kane
#
# Kevin C. O'Kane
# anamfianna@earthlink.net
# okane@cs.uni.edu
#
#
# This program is free software; you can redistribute it and/or modify
# it under the terms of the GNU General Public License as published by
# the Free Software Foundation; either version 2 of the License, or
# (at your option) any later version.
#
# This program is distributed in the hope that it will be useful,
# but WITHOUT ANY WARRANTY; without even the implied warranty of
# MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the
# GNU General Public License for more details.
#
# You should have received a copy of the GNU General Public License
# along with this program; if not, write to the Free Software
# Foundation, Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
#
# ++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
# tab.mps January 2, 2008
set lev(0)="0000\Browser Tree"
set ord=1000
for Do
. read cmd
. if '$Test break
. if cmd="" continue
. for i=0:1:10 if $extract(cmd,i+1,i+1)'="." quit
. for do
.. if $extract(cmd,1,1)="."!($extract(cmd,1,1)=" ") set cmd=$extract(cmd,2,512) quit
.. break
. set lev(i)=cmd
. read order // line2
. for do
.. if $extract(order,1,1)="."!($extract(order,1,1)=" ") set order=$extract(order,2,512) quit
.. break
. set x=""
. for j=0:1:i-1 set x=x_""""_lev(j)_""","
. set x=x_""""_lev(i)_""""
. set x="^lib("_x
. set %=x_")"
. set order="@"_order
. set @%=order
. write %," -> ",order,!
halt
|
The results for OSU Medline are here:
http://www.cs.uni.edu/~okane/source/ISR/medline.tab.gz.
Finally, once the internal hierarchy is built,
the program that displays the indices is here:
|
#!/usr/bin/mumps
#++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
#+
#+ Mumps Web Software Library
#+ Copyright (C) 2000, 2005, 2006, 2008 by Kevin C. O'Kane
#+
#+ Kevin C. O'Kane
#+ anamfianna@earthlink.net
#+ okane@cs.uni.edu
#+
#+
#+ This program is free software; you can redistribute it and/or modify
#+ it under the terms of the GNU General Public License as published by
#+ the Free Software Foundation; either version 2 of the License, or
#+ (at your option) any later version.
#+
#+ This program is distributed in the hope that it will be useful,
#+ but WITHOUT ANY WARRANTY; without even the implied warranty of
#+ MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the
#+ GNU General Public License for more details.
#+
#+ You should have received a copy of the GNU General Public License
#+ along with this program; if not, write to the Free Software
#+ Foundation, Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
#+
#++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
# index.mps January 2, 2008
znumber
html Content-type: text/html &!&!<html>
html <head><title>Mumps Web Folders</title></head>
html <body vlink=darkblue link=darkblue bgcolor=silver>&!
html <!-- Mumps Web Folders, Copyright (C) 2001 Kevin C. O'Kane &!
html Mumps Web Folders comes with ABSOLUTELY NO WARRANTY; &!
html This is free software, and you are welcome to redistribute it&!
html under certain conditions as set forth in the GNU GPL -->&!
if '$data(^count(1)) set ^count(1)=1 set ^date(1)=$zd set date=$zd
else set ^count(1)=^count(1)+1 set date=^date(1)
if '$data(ml) set ml=1
set line=0
if '$data(array) set array="lib"
set %1="<a href=""index.cgi?array="_array_"&ml="
set %2="<a href="
set cls="<img align=top src=""/folder03.gif"" border=0>"
set lev=1,bx=0,ep=0,c(0)=""
for i=0:1:20 do
. set b(i)=""
. set x="a"_i
. set a(i)=""
. if $data(@x) set b(i)=@x,ep=i
do layer
html <center>
html </body></html>
halt
layer set x(lev)="^"_array_"("
set AFLG=0
for i=1:1:lev do
. set x(lev)=x(lev)_"a("_i_")"
. if i'=lev set x(lev)=x(lev)_","
set x(lev)=x(lev)_")"
set a(lev)=-1
if b(lev)'="",lev<ml do
. set a(lev)=""
a1 set a(lev)=$order(@x(lev))
if a(lev)="" set lev=lev-1 quit
set p="array="_array
if lev>bx do parm1
set n="<br>",nn=""
for i=1:1:lev-1 set n=n_"..."
for i=1:1:lev-1 set nn=nn_"..."
if $data(@x(lev))>1,ml>lev,b(lev)=a(lev) do
. html </a><br><a name=node&~lev~></a>
. write nn," ",%1,lev set line=0
. do parm
. Html ">
. Html <img align=middle src="/folder05.gif" border=0>
. set line=line+1
. set AFLG=1
. goto a3a
if $data(@x(lev))=1,lev=ml do
. write n
. set n=""
. goto a3a
if $data(@x(lev))'=1 do
. if line>25 do
.. write "<br>" set line=0
.. write nn,%1,lev+1
. else write n,%1,lev+1
. do parm
. write """>",cls set line=line+1
. set AFLG=1
a3a if AFLG do tok
if 'AFLG do
. if $extract(@x(lev),1,1)="@" do
.. set p=$extract(@x(lev),2,100)
. write %2,p
. html >
. if $data(@x(lev))=1 set line=line+1 html &~n~&nbsp;<img src=/blueball.gif border=0 align=top>&nbsp;
. do tok
. Html </a>
if lev<ml do
. if $data(@x(lev))>1,b(lev)=a(lev) do
.. if $extract(@x(lev),1,1)="@" do
... set p=$extract(@x(lev),2,100)
.. write %2,p set line=line+1
.. Html >
.. Html </a>&!
if @x(lev)="" goto a2a
set x=@x(lev)
if lev<ml goto a2a ; print only low level
if $extract(x,1,1)="@" do
. set x=$extract(x,2,255)
a2a if lev>bx do parm1
if $data(@x(lev))>1,ml>lev,b(lev)=a(lev) do
. set lev=lev+1
. do layer
. goto a1
goto a1
parm for i=1:1:lev quit:a(i)=-1 write "&a",i,"=",$zh(a(i))
html #node&~lev~
quit
# tok write !,$piece(a(lev)," ",2,99)
tok write !,a(lev)
if AFLG html </a>
quit
parm1 set bx=lev
for i=1:1:lev set c(i)=a(i)
quit
|
Which produces an initial browser display such as the following:
Upon clicking on the letter "o", the display becomes:
Upon clicking on the "obstructive" link, the display becomes:
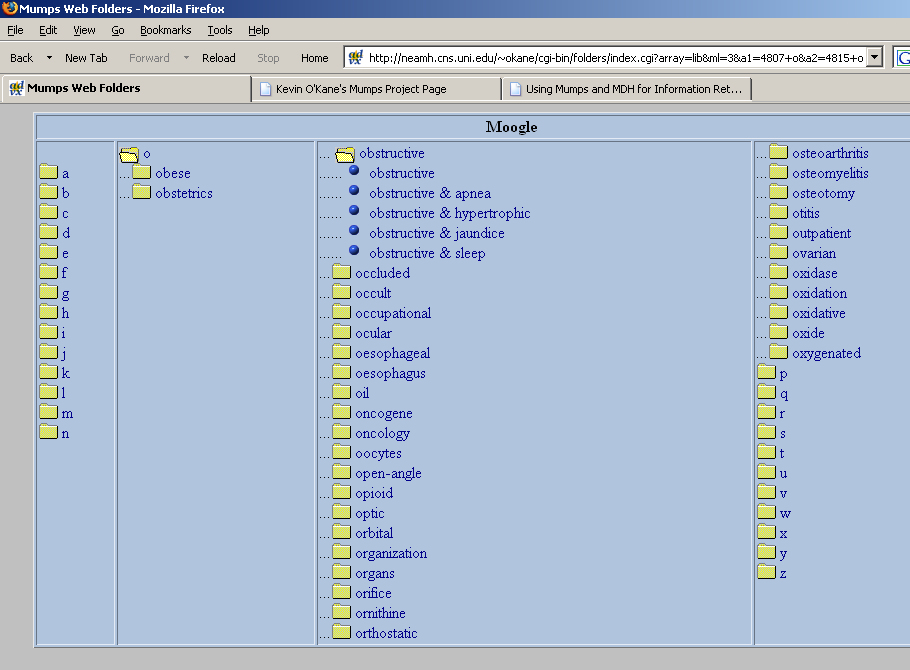
Upon clicking the button "obstructive & apnea" the webFinder
program is invoked with the keys selected (and'ed) and the display
becomes:
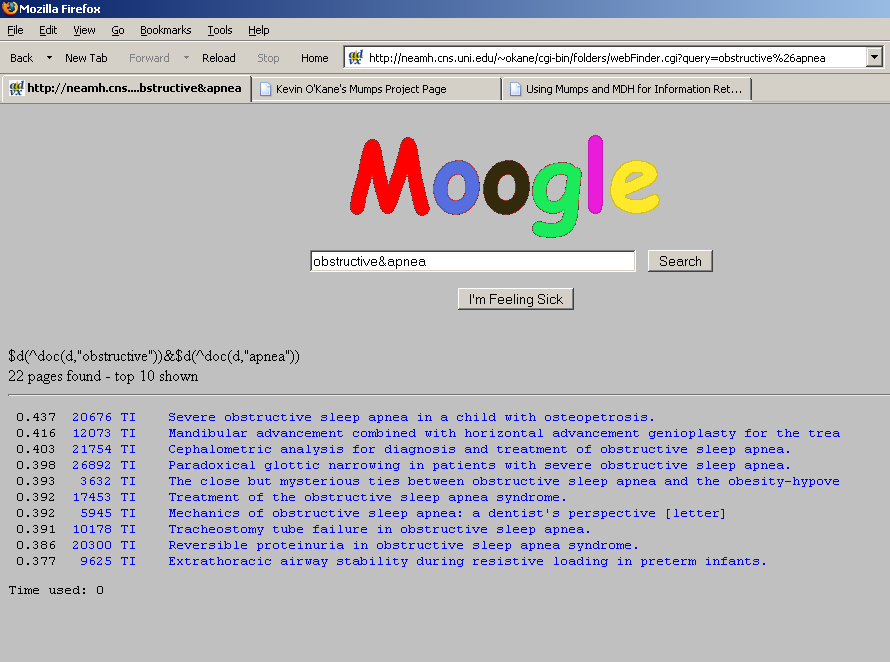
If the user clicks the first retrieved title, the program
display.mps is run:
|
#!/usr/bin/mumps
#+++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
#+
#+ Mumps ISR Software Library
#+ Copyright (C) 2005, 2006, 2008 by Kevin C. O'Kane
#+
#+ Kevin C. O'Kane
#+ okane@cs.uni.edu
#+
#+
#+ This program is free software; you can redistribute it and/or modify
#+ it under the terms of the GNU General Public License as published by
#+ the Free Software Foundation; either version 2 of the License, or
#+ (at your option) any later version.
#+
#+ This program is distributed in the hope that it will be useful,
#+ but WITHOUT ANY WARRANTY; without even the implied warranty of
#+ MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the
#+ GNU General Public License for more details.
#+
#+ You should have received a copy of the GNU General Public License
#+ along with this program; if not, write to the Free Software
#+ Foundation, Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
#+
#++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
# display.mps March 23, 2008
html Content-type: text/html &!&!
html <html><body bgcolor=silver>
if '$data(ref) write "Missing reference number </body></html>" halt
open 1:"osu.medline,old"
if '$test write "osu.medline not found",! halt
use 1 set %=$zseek(^doc(ref))
html <table bgcolor=lightsteelblue align=center width=60% border><tr><td><i>Mesh Headings:</i><br>
set flg=0
for do
. use 1
. read a // find the title
. if a="" do
.. html No abstract available
.. html </table></body></html>
.. halt
. if $extract(a,1,3)="MH " do
.. use 5
.. set i=$find(a,"/")
.. if i=0 set i=255
.. else set i=i-2
.. set a=$extract(a,7,i)
.. do $zwi(a)
.. for ww=$zwp do
... if ww?.p continue
... set wx=ww
... if $data(^lib($e(wx,1,1),wx)) do
.... html <a href=index.cgi?
.... html array=lib&ml=3&a1=&~$e(wx,1,1)~&a2=&~wx~#node2>
.... write ww,"</a> "
... else write ww," "
. if $extract(a,1,3)="TI " do
.. use 5
.. do $zwi($extract(a,7,255))
.. html <tr><td><i>Title:&nbsp;</i>
.. for ww=$zwp do
... set wx=ww
... if $data(^lib($e(wx,1,1),wx)) do
.... html <a href=index.cgi?
.... html array=lib&ml=3&a1=&~$e(wx,1,1)~&a2=&~wx~#node2>
.... write ww,"</a> "
... else write ww," "
.. html </td><tr><td><i>Abstract:</i>&nbsp;
. if $extract(a,1,3)="AB " for do
.. use 5
.. do $zwi($extract(a,7,255))
.. for ww=$zwp do
... set wx=ww
... if $data(^lib($e(wx,1,1),wx)) do
.... html <a href=index.cgi?
.... html array=lib&ml=3&a1=&~$e(wx,1,1)~&a2=&~wx~#node2>
.... write ww,"</a> "
... else write ww," "
.. use 1 read a
.. if a=""!($extract(a,1,3)'=" ") set flg=1 break
. if flg break
use 5
html <tr><td><i>Related Documents</i>:
set d=""
for do
. set d=$order(^dd(ref,d))
. if d="" break
. use 1 set %=$zseek(^doc(d))
. for do
.. use 1
.. read a // find the title
.. if a="" do
... html </table></body></html>
... halt
.. if $extract(a,1,3)="TI " do
... use 5
... html <br><a href=display.cgi?ref=
... write d,">",d,"</a> "
... do $zwi($extract(a,7,255))
... for ww=$zwp do
.... set wx=ww
.... if $data(^lib($e(wx,1,1),wx)) do
..... html <a href=index.cgi?
..... html array=lib&ml=3&a1=&~$e(wx,1,1)~&a2=&~wx~#node2>
..... write ww,"</a> "
.... else write ww," "
html </table></body></html>
|
The program above accesses the original source text from
the OSU Medline data base. In an earlier step,
the program sourceOffsets.mps was run
to extract the offsets of the original text.
and the display becomes:
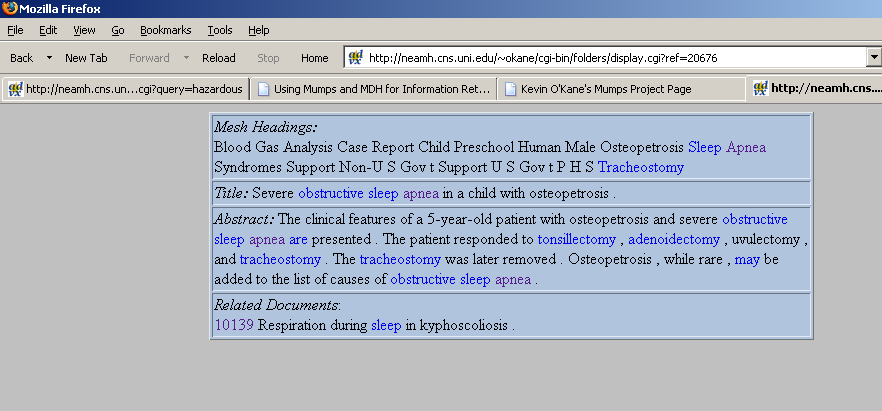
The web index program is capable of multiple levels
of hierarchy but the combinatorics of word
combinations
become very large unless care is taken only to display very
common, broadly appearing terms at the higher levels.
In the above display, if the user clicks on a a highlighted word,
they are taken to the initial display of folders with the
folder for the keyword clicked open. Also listed at the bottom
are the article numbers of related documents (from the doc-doc
matrix). Clicking on one of the document numbers will display
the selected document with its keywords highlighted.
An online demo of the above programs is
available
here (depending on server availability).
Note: this operates on a student server and may be down or
operate very slowly at times.
Indexing Text Features in Genomic Repositories
Since the widespread adoption of the Internet in the
early 1990's, there has been explosive growth in
machine readable data bases both in the form of
online data bases as well as web page based
content. Readily accessible indexable
content now easily ranges into terabytes.
When indexing very large data sets, special procedures
should be used to maximize efficiency.
The following is a case study based on indexing the text content
of genomic data bases.
Since 1990, the massive growth in genetic and protein databases has
created a pressing need for tools to manage, retrieve and analyze the information contained
in these libraries. Traditional tools to organize, classify and extract information have
often proved inadequate when confronted with the overwhelming size and density of
information which includes not only sequence and structural data, but also text that describes
the data's origin, location, species, tissue sample, journal articles, and so forth. As of this
writing, the NCBI (National Center for Biotechnology Information, part of the National Institutes
of Health) GenBank library alone consists of nearly 84 billion bytes of data and it is only one of
several data banks storing similar information. The scope and size of these databases continues to rapidly grow
and will continue to do so for many years to come as will the demand for access.
A typical entry in GenBank looks like:
(from: ftp://ftp.ncbi.nih.gov/genbank/ )
|
Web Abstract Display Program
|
LOCUS AAB2MCG2 1276 bp DNA linear PRI 23-AUG-2002
DEFINITION Aotus azarai beta-2-microglobulin precursor exons 2, 3, and
complete cds.
ACCESSION AF032093 AF032094
VERSION AF032093.1 GI:3287308
KEYWORDS .
SEGMENT 2 of 2
SOURCE Aotus azarai (Azara's night monkey)
ORGANISM Aotus azarai
Eukaryota; Metazoa; Chordata; Craniata; Vertebrata; Euteleostomi;
Mammalia; Eutheria; Primates; Platyrrhini; Cebidae; Aotinae; Aotus.
REFERENCE 1 (bases 1 to 1276)
AUTHORS Canavez,F.C., Ladasky,J.J., Muniz,J.A., Seuanez,H.N., Parham,P. and
Cavanez,C.
TITLE beta2-Microglobulin in neotropical primates (Platyrrhini)
JOURNAL Immunogenetics 48 (2), 133-140 (1998)
MEDLINE 98298008
PUBMED 9634477
REFERENCE 2 (bases 1 to 1276)
AUTHORS Canavez,F.C., Ladasky,J.J., Seuanez,H.N. and Parham,P.
TITLE Direct Submission
JOURNAL Submitted (31-OCT-1997) Structural Biology, Stanford University,
Fairchild Building Campus West Dr. Room D-100, Stanford, CA
94305-5126, USA
COMMENT On or before Jul 2, 1998 this sequence version replaced gi:3265029,
gi:3265028.
FEATURES Location/Qualifiers
source 1..1276
/organism="Aotus azarai"
/mol_type="genomic DNA"
/db_xref="taxon:30591"
mRNA join(AF032092.1:<134..200,66..344,1023..>1050)
/product="beta-2-microglobulin precursor"
CDS join(AF032092.1:134..200,66..344,1023..1036)
/codon_start=1
/product="beta-2-microglobulin precursor"
/protein_id="AAC52107.1"
/db_xref="GI:3289965"
/translation="MARFVVVALLVLLSLSGLEAIQRXPKIQVYSRHPAENGKPNFLN
CYVSGFHPSDIEVDLLKNGKKIEKVEHSDLSFSKDWSFYLLYYTEFTPNEKDEYACRV
SHVTLSTPKTVKWDRNM"
mat_peptide join(AF032092.1:194..200,66..344,1023..1033)
/product="beta-2-microglobulin"
intron <1..65
/number=1
variation 3
/note="allele 1"
/replace="g"
exon 66..344
/number=2
intron 345..1022
/number=2
exon 1023..1050
/number=3
intron 1051..>1276
/number=3
ORIGIN
1 caagttatcc gtaattgaaa taccctggta attaatattc atttgtcttt tcctgatttt
61 ttcaggtrct ccaaagattc aggtttactc acgtcatccg gcagagaatg gaaagccaaa
121 ttttctgaat tgctatgtgt ctgggtttca tccgtccgac attgaagttg acttactgaa
181 gaatggaaag aaaattgaaa aagtggagca ttcagacttg tctttcagca aggactggtc
241 tttctatctc ttgtactaca ccgagtttac ccccaatgaa aaagatgagt atgcctgccg
301 tgtgagccat gtgactttat caacacccaa gacagtaaag tggggtaagt cttacgttct
361 tttgtaggct gctgaaagtt gtgtatgggt agtcatgtca taaagctgct ttgatataaa
421 aaaaattcgt ctatggccat actgccctga atgagtccca tcccgtctga taaaaaaaaa
481 tcttcatatt gggattgtca gggaatgtgc ttaaagatca gattagagac aacggctgag
541 agagcgctgc acagcattct tctgaaccag cagtttccct gcagctgagc agggagcagc
601 agcagcagtt gcacaaatac atatgcactc ctaacacttc ttacctactg acttcctcag
661 ctttcgtggc agctttaggt atatttagca ctaatgaaca tcaggaaggt ataggccttt
721 ctttgtaaat ccttctatcc tagcatccta taatcctgga ctcctccagt actctctggc
781 tggattggta tctgaggcta gtaggtgggg cttgttcctg ctgggtagct ccaaacaagg
841 tattcatgga taggaacagc agcctatttt gccagcctta tttcttaata gttttagaaa
901 tctgttagta cgtggtgttt tttgttttgt tttgttttaa cacagtgtaa acaaaaagta
961 catgtatttt aaaagtaaaa cttaatgtct tcctttttct ttctccactg tctttttcat
1021 agatcgaaac atgtaaccag catcatggag gtaagttctt gaccttaatt aaatgttttt
1081 tgtttcactg gggactattt atagacagcc ctaacatgat aaccctcact atgtggagaa
1141 cattgacaga gtagcatttt agcaggcaaa gaggaatcct atagggttac attccctttt
1201 cctgtggagt ggcatgaaaa aggtatgtgg ccccagctgt ggccacatta ctgactctac
1261 agggagggca aaggaa
|
An annotated example GenBank record is available
here
while a complete description of the NCBI data base can be found
here.
Currently, retrieval of genomic data is mainly based on well-established programs such as FASTA
(Pearson, 2000, see: ) and BLAST (Altschul, 1997), that match candidate nucleotide sequences against massive
libraries of sequence acquisitions. There have been few efforts to provide access to genomic data keyed
to the extensive text annotations commonly found in these data sets. Among the few systems that deal
with keyword based searching are the proprietary SRS system (Thure and Argos, 1993a, 1993b) and
PIR (Protein Information Resource) (Wu 2003). These are limited, controlled vocabulary systems whose
keys are from manually prepared annotations. To date, there have been no systems reported to directly
generate indices from the genomic data sets themselves. The reasons for this are several: the very
large size of the underlying data sets, the size of intermediate indexing files, the complexity of the
data, and the time required to perform the indexing.
The system described here, MARBL (Mumps Analysis and Retrieval from Bioinformatics Libraries), is an
application to integrate multiple, very large, genomic, databases into a unified
data repository through open-source components; and to provide fast, web-based keyword based access
to the contents.
Sequences retrieved by MARBL can be post-processed by FASTA (Pearson, 2000), Smith-Waterman (Smith, 1981)
and elements of EMBOSS (the European Molecular Biology Open Software Suite). While FASTA and, especially,
Smith-Waterman, are more sensitive (Shpaer et al. 1996) than BLAST, they are also more time consuming.
However, by first extracting from the larger database a subset of candidate accessions, the number of
sequences to be aligned by these algorithms can be reduced significantly with corresponding
reduction in the overall processing time.
Implementation
Most genomic databases include, in addition to nucleotide and protein sequences, a wealth of text
information in the form of descriptions, keywords, annotations, hyper-links to text articles, journals
and so forth. In many cases, the text attachments to the data are greater in size that the actual
sequence data. Identifying the important keyword terms from this data and assigning a relative weight
to these terms is one of the problems addressed in this system.
While indexing can be approached from the perspective of assignment to pre-existing categories and
hierarchies such as the National Library of Medicine MeSH (Medical Subject Headings) (Hazel, 1997),
derivative indexing is better able to adapt to changes in a rapidly evolving discipline as the terms
are dynamically extracted directly from the source material rather than waiting for manual analysis. Existing
keyword based genomic retrieval systems are primarily based on assignment indexing whereas the approach
taken here is based on derivative indexing, where both queries and documents are encoded into a common
intermediate representation and metrics are developed to calculate the coefficients of similarity between queries
and documents. Documents are ranked according to their computed similarity to the query and presented to the
user in rank order. Several systems employing this and related models have been implemented such as
Smart (Salton, 1968, 1971, 1983, 1988), Instruct (Wade, 1988), Cansearch (Pollitt, 1987) and
Plexus (Vickery, 1987a, 1987b). More recently, these approaches have been used to index Internet web pages and provide collaborative filtered recommendations regarding similar texts to book buyers at Amazon.com (Linden, 2003).
In this system, genomic accessions are represented by vectors that reflect accession content through descriptors
derived from the source text by analysis of word usage (Salton,1968, 1971, 1983, 1988; Willett,
1985; Crouch, 1988). This approach can be further enhanced by identifying clusters of similar documents
(El-Hamdouchi et al.,1988, 1989). Similarly, term-term co-occurrence matrices can be constructed to
identify similar or related terms and these can be automatically included into queries to enhance
recall or to identify term clusters. Other techniques based on terms and queries have
also been explored (Salton, 1988; Williams, 1983).
The vector model is rooted in the construction of document vectors consisting of the weights of
each term in each document. Taken collectively, the document vectors constitute a document-term matrix
whose rows are document vectors. A document-term matrix can have millions of rows, more than 22 million
in GenBank case, and thousands of columns (terms), more than 500,000 in GenBank. This yields a matrix with
potentially trillions of possible elements which must be quickly addressable not by numeric indices but by
text keys. Additionally, to enhance information retrieval speed, an inverted matrix of the same size is needed which
doubles the overall storage requirements. Fortunately, however, both matrices are very sparse.
Given the nature of the problem, namely, manipulating massive, character string indexed sparse
matrices, we implemented the system in Mumps.
Using Mumps global arrays, an accession-term matrix appears in the Mumps language as an array
of the form: ^D(Accession,Term) where both Accession and Term are text strings. The matrix is indexed
row-wise by accession codes and column-wise by text derived terms. This approach vastly simplifies
implementation of the basic information storage and retrieval model. For example, the main Mumps indexing program used
in the basic protocol described below is about 76 lines of code (excluding in-line C functions).
The highly concise nature of the Mumps language permits rapid deployment and minimizes maintenance problems that
would be the case with more complex coding systems.
FASTA (Pearson, 2000) and Smith-Waterman (Smith and Waterman, 1981) are sequence alignment procedures to
match candidate protein or NA sequences to entries in a database. Sequences retrieved as a result of text searches
with the system described here can be post-processed by FASTA and the Smith-Waterman. Of these,
the Smith-Waterman algorithm is especially sensitive and accurate but also relatively time consuming. Using this system to
isolate candidate sequences by text keywords and subsequently processing the resulting subset of the larger database, results in considerable
time savings. In our experiments we used the Smith-Waterman program available as part of the FASTA package
developed by W. R. Pearson (Pearson 2000). Additionally, the output of this system is compatible
with the many genomic analysis programs found in the open source EMBOSS collection.
The system software is compatible with several genomic database input formats, subject to preprocessing by filters. In the
example presented here, the NCBI GenBank collection was used. GenBank consists of accessions which contain sequence and related
data collected by researchers throughout the world.
Two protocols were developed to index the GenBank data sets.
Initially, we tried a direct, single step, vector space model protocol that constructed the
accession-term matrix directly from the GenBank files. However, experiments revealed that this approach was unacceptably slow
when used with large data sets. This resulted in the development of a multi-step
protocol that performed the same basic functions but as a series of steps designed to improve
overall processing speed. The discussion below centers on the multi-step protocol although
timing results are given for both models. The work was performed on a Linux based,
dual processor hyper-threaded Pentium Xeon 2.20 GHz system with 1 GB of main memory and
dual 120 GB EIDE 7,200 rpm disk drives.
The entire GenBank data collection consisted of approximately 83.5 GB of data at the time of these experiments.
When working with data sets of smaller size, relatively straightforward approaches to text processing can
be used with confidence. However, when working with data sets of very large dimensions, it soon became
apparent that special strategies would be needed in order to reduce the complexity of the processing
problem. During indexing, as the B-tree grew, delays due to I/O became significant when the size of
the data set exceeded the amount of physical memory available. At that point, the memory I/O cache
became inadequate to service I/O requests without significant actual movement of data to and from external
media. When this happened, CPU utilization was observed to drop to very low values while input/output
activity grew to system maximum. Once page thrashing began, overall progress to index the data set
slowed dramatically.
In order to avoid this problem, the multi-step protocol was devised in which the indexing tasks were
divided into multiple steps and sort routines were employed in order to prepare intermediate data
files so that at each stage where the database was being loaded into the B-tree, the keys were
presented to the system in ascending key order thus inducing an effectively sequential data set build
and eliminating page thrashing. While this process produced a significant number of large intermediate
files, it was substantially faster than unordered key insertion.
Data Sets
The main data sets used were from the NCBI GenBank collection (ftp://ftp.ncbi.nlm.nih.gov):
-
The GenBank short directory, gbsdr.txt consisting of locus codes
which, at this writing, is approximately 1.45 billion bytes in
length and has approximately 18.2 million entries.
-
The nucleotide data, the accessions, are stored in over 300 gzip compressed files.
Each file is about 220 megabytes long and consists of nucleotide accessions. We pre-process
each file with a filter program that extracts text and other information. Pre-processing
results in a format similar to the EMBL (European Micro Biology Laboratory) format and this
makes for faster processing in subsequent steps as well as greatly reducing disk storage requirements.
For example, the file gbbct1.seq, currently 250,009,587 bytes in length,
reduced to 8,368,295 bytes after pre-processing.
-
Optionally, a list of NCBI manually derived multi-word keys from the file gbkey.idx
(502,549,211 bytes). Processing of these keys is similar to that of derived keys but only a default,
minimum weight is produced.
-
In addition to text found in the accessions, GenBank, as well as many other data resources,
contains links to on-line libraries of journal articles, books, abstract and so forth.
These links provided additional sources of text keys related to the
accessions in the originating database.
Multiple Step Protocol
The multiple step protocol, shown in Figure 1, separated the work into several steps and was based on the observation
that using system sort facilities to preprocess data files resulted in much faster database
creation since the keys can be loaded into the B-tree database in ascending key order. This
observation was founded in an early experiment in which an accession-term matrix was constructed by
loading the keys from a 5 million accessions file sorted by ascending accession key. The load
procedure itself used a total of 1,032 seconds (17.2 minutes). On the other hand, loading the
keys directly from a file not sorted by accession was 7.1 times slower requiring
7,333 seconds (122.2 minutes) to load.
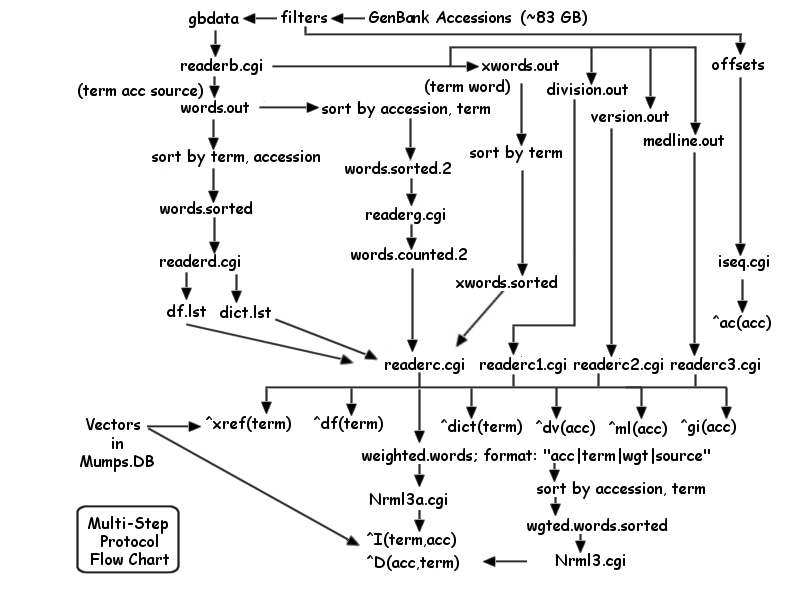
Figure 1
The main text analysis procedure reads the filtered version of the accession files. Lines of text were
scanned, punctuation and extraneous characters were removed, words matching entries in the stop list were
discarded, and, finally, words were processed to remove suffixes and consolidated into groups based on word
stems (readerb.cgi). Each term was written to an output file (words.out) along with its accession code and
a code indicating the source of the data. A second file was also produced that associated each processed
stem along with the original form of the term (xwords.out). The output files were sorted
concurrently. The xwords.out file was sorted by term with duplicate entries discarded while the words.out
file was sorted to two output files: words.sorted, ordered by term then accession code, and
words.sorted.2 ordered by accession code then term.
The file words.sorted was processed to count word usage (readerd.cgi). As the file was ordered
by term then accession code, multiple occurrences of a word in a document appear on successive lines.
The program deleted words whose overall frequency of occurrence was too low or high. Files df.lst and
dict.lst were produced which contain, respectively, for each term, the number of accessions in which
it appears in, and the total number of occurrences.
The file words.sorted.2 (sorted by accession code and term) was processed by readerg.cgi to produce
words.counted.2 which generated for each accession, the number of times each term occurred in an
accession and a string of code letters giving the original sources of the term (from the original input
line codes). This file was ordered by accession and term.
The files xwords.sorted, df.lst, dict.lst and words.counted.2 were processed by readerc.cgi to produce
internal data vectors and an output file named weighted.words which contained the accession code, term,
the calculated inverse document frequency weight of the term, and source code(s) for the term.
If the calculated weight of a term in an accession was below a threshold, it was discarded. Since
the input file words.counted.2 was ordered by accession then by term, the output file weighted.words was
also ordered by accession then term.
Finally, the Nrml3a.cgi constructed the term-accession matrix (^I) from the term sorted file weighted.words
and Nrml3.cgi built the accession-term matrix (^D) from wgted.words.sorted which was ordered by accession
and term. In this final step, the database assumed its full size and it was this step that is most
critical in terms of time. As each of the matrices were ordered according to their first, then second
indices, the B-tree was built in ascending key order.
Retrieval
Retrieval is via a web page interface or an interactive keyboard based program. Queries are expressed
as logical expressions of terms, possibly including wildcards. Queries may be restricted to data particular
sources (title, locus, etc.) or specific divisions (ROD, PRI, etc). When a query is submitted to the
system it is first expanded to include related terms for any wildcards. The expression is converted into
a Mumps expression and candidate accession codes of those accessions containing terms from the query
are identified. The Mumps expression is applied to each identified candidate accession. A similarity
coefficient between the accession and the query is calculated based on the weight of the terms in the
accessions using a simple similarity formula.
From the accessions retrieved, the user can view the original NCBI accession page, save the accession
list for further reference or convert the accessions to FASTA format and match the accessions against
a candidate sequence with the FASTA or the Smith-Waterman algorithm. By means of the GI code from the
VERSION field in the original GenBank accession, a user can access full data concerning the retrieved accession
directly from NCBI. Also stored is the Medline access code which provides direct entry into the Medline database for
the accession.
Retrieval times are proportional to the amount of material retrieved, the complexity of the query, and the
number of accessions in which each query term appears.. For specific queries that retrieve only a few
accessions, processing times less than 1 second are typical.
Results and Discussion
Some overall processing statistics for the two protocols are given in Table 1. As can be seen,
the multi-step protocol performed significantly better than the basic protocol.
|
Table 1 - Processing Time Statistics (in minutes)
|
|
Accessions Processed | 1,000,000 | 5,000,000 | 22,318,882
|
|
Multi-Step Protocol | 63.9 | 350.8 | 2,016.1
|
|
Basic Protocol | 246.9 | 1,735.61 | 6994.7
|
The dimensions of the final matrices are potentially of vast size: 22.3 million by
501,614 in this case. Potentially, this implies a matrix of 11.5 trillion elements. However,
the matrix is very sparse and the file system stores only those elements which actually exist.
After processing the entire GenBank, the actual database was only 23 GB although at its largest,
before compaction of unused space, it reached 44 GB.
Evaluation of information retrieval effectiveness from a data set of this size is clearly difficult as there are
few benchmarks against which to compare the results. However, NCBI distributes a file of keyword
phrases with GenBank gbkey.idx (502,549,211 bytes). This file contains submission author assigned
keyword phrases and associated accession identifiers. Of the 48,023 unique keys in gbkey.idx
(after removal of special characters and words less than three characters in length), 26,814 keys
were the same as the keys selected by MARBL. The 21,209 keys that differed were, for the most
part, words of very high or low frequency that the system rejected due to preset thresholds.
Alternatively, the MARBL system identified and retained a highly specific 501,614 terms, many
of which were specific codes used to identify genes.
When comparing the accessions linked to keywords in gbkey.idx with MARBL derived accessions, it
was clear that MARBL discovered vastly more linkages than the NCBI file identified. For example,
the keyword zyxin (the last entry in gbkey.idx) was linked to 4 accessions by gbkey.idx but
MARBL detected 336 accessions. In twelve other queries based on terms randomly selected from gbkey.idx,
MARBL found more accessions than were listed in gbkey.idx in nine cases and the same number
in three cases. On average, each MARBL derived keyword points to 130.34 accessions whereas gbkey.idx
keys, on average, points to 6.80 accessions.
We compared MARBL with BLAST by entering the nucleotide sequence of a Bacillus anthracis bacteriophage that
was of interest to a local researcher. BLAST retrieved 24 accessions, with one scoring 1,356, versus the
next highest with a score of 50. The highest scoring accession was the correct answer, while the
remainder were noise. When we entered the phrase anthracis & bacteriophage to the MARBL information retrieval
package, only one accession was retrieved, the same one that received the highest score from
BLAST. BLAST took 29 seconds, MARBL information retrieval took 10 seconds. It should be noted,
however, that BLAST searches are not based on keywords but on genomic sequences.
Mumps is an excellent text indexing implementation language (O'Kane, 1992). Mumps programs
are concise and are easily maintained and modified. The string indexed global arrays,
underpinned by the effectively unlimited file sizes supported by the BDB, make it possible
to design very large, efficient systems with minimal effort. In all, there were 10 main
indexing routines with a total of 930 lines of Mumps code (including comments) for
an average of 93 lines of code per module. On the other hand, the C programs generated by
the Mumps compiler amounted to 21,146 lines of code, not counting many thousands of lines in
run-time support and database routines. The size of the C routines is comparable to reported code
sizes for other information storage and retrieval projects such as Wade (1988) who reports that Instruct as
approximately 6,000 lines of Pascal code, and Plexus (Vickery and Brooks, 1987a) reported as
approximately 10,000 lines although, due to differences in features, these figures should not be
used for direct comparisons.
An example of this system in its current state can be seen
here.
N-gram encoding (Salton83, pages 93-94)
During World War II, n-grams, fixed length consecutive series of "n" characters, were developed by
cryptographers to break substitution ciphers. Applying n-grams to indexing, the text, stripped of
non-alphabetic characters, is treated as a continuous stream of data that is segmented into
non-overlapping fixed length words. These words can then form the basis of the indexing vocabulary.
In the following experiment, the OSU TREC9 data base,
is read and the text reduced to non-overlapping 3 letter n-grams.
First, the input text is pre-processed to remove non-alpha characters and converted to
lower case. The result is written in a
FASTA format consisting of a title line beginning with a ">" followed by the title
of the article followed by
a long single line of text comprising the text and body of the article
converted as noted above. (Note: if the lines of text exceed 2,500 characters, the
Mumps Compiler configure parameter --with-strmax=val parameter will need to be increased
from its default value of 2500.)
|
Conversion of TREC9 Data Base to FASTA Format
|
#+++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
#+
#+ Mumps ISR Software Library
#+ Copyright (C) 2007 by Kevin C. O'Kane
#+
#+ Kevin C. O'Kane
#+ okane@cs.uni.edu
#+
#+
#+ This program is free software; you can redistribute it and/or modify
#+ it under the terms of the GNU General Public License as published by
#+ the Free Software Foundation; either version 2 of the License, or
#+ (at your option) any later version.
#+
#+ This program is distributed in the hope that it will be useful,
#+ but WITHOUT ANY WARRANTY; without even the implied warranty of
#+ MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the
#+ GNU General Public License for more details.
#+
#+ You should have received a copy of the GNU General Public License
#+ along with this program; if not, write to the Free Software
#+ Foundation, Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
#+
#++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
# fasta.mps April 1, 2007
open 1:"osu.medline,old"
if '$test write "osu.medline file not found",! halt
set f=0
for do
. use 1
. read line
. if '$test halt
. if $e(line,1,2)="TI" use 5 write:f ! write "> ",$e(line,7,256),!,$$cvt(line) set f=1 quit
. if $e(line,1,2)'="AB" quit
. use 5 write $$cvt(line)
. for do // for each line of the abstract
.. use 1 read line
.. if '$test use 5 write ! halt // no more input
.. if line="" break
.. use 5 write $$cvt(line)
. use 5 write !
. set f=0
halt
cvt(line)
set buf=""
for i=7:1:$l(line) do
. if $e(line,i)?1A set buf=buf_$e(line,i)
set buf=$zlower(buf)
quit buf
|
A substantially faster version, written in C:
|
Conversion of TREC9 Data Base to FASTA Format
|
// #+++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
// #+
// #+ Mumps ISR Software Library
// #+ Copyright (C) 2007 by Kevin C. O'Kane
// #+
// #+ Kevin C. O'Kane
// #+ okane@cs.uni.edu
// #+
// #+
// #+ This program is free software; you can redistribute it and/or modify
// #+ it under the terms of the GNU General Public License as published by
// #+ the Free Software Foundation; either version 2 of the License, or
// #+ (at your option) any later version.
// #+
// #+ This program is distributed in the hope that it will be useful,
// #+ but WITHOUT ANY WARRANTY; without even the implied warranty of
// #+ MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the
// #+ GNU General Public License for more details.
// #+
// #+ You should have received a copy of the GNU General Public License
// #+ along with this program; if not, write to the Free Software
// #+ Foundation, Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
// #+
// #++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
//
// # MDH-fasta.cpp April 1, 2007
#include <stdio.h>
#include <stdlib.h>
#include <ctype.h>
#include <assert.h>
#include <string.h>
void cvt(char *line, char *buf) {
int i,j;
buf[0] = '\0';
j = 0;
for (i=6; line[i] != '\0'; i++)
if (isalpha(line[i])) buf[j++] = tolower(line[i]);
buf[j] = 0;
}
int main () {
FILE *u1;
char line[512],buf[8192];
int i,j,f;
u1 = fopen("osu.medline","r");
assert (u1 != NULL);
f=0;
while (1) {
if (fgets(line, 512, u1) == NULL) break;
if (strncmp(line,"TI",2) == 0) {
if (f) printf("\n");
printf("> %s",&line[6]);
cvt (line, buf);
printf("%s",buf);
f=1;
continue;
}
if (strncmp(line,"AB",2) != 0) continue;
cvt(line,buf);
printf("%s",buf);
while (1) {
if (fgets(line, 512, u1) == NULL) break;
if (strlen(line) == 1) break;
cvt(line,buf);
printf("%s",buf);
}
printf("\n");
f=0;
}
return EXIT_SUCCESS;
}
|
The results look like the following:
|
TREC9 Data Base in FASTA Format Example
|
> The binding of acetaldehyde to the active site of ribonuclease: alterations in catalytic activity and effects of phosphate.
thebindingofacetaldehydetotheactivesiteofribonucleasealterationsincatalyticactivityandeffectsofphosphateribonucleaseawasreacte
dwithccacetaldehydeandsodiumcyanoborohydrideinthepresenceorabsenceofmphosphateafterseveralhoursofincubationatdegreescphstablea
cetaldehydernaseadductswereformedandtheextentoftheirformationwassimilarregardlessofthepresenceofphosphatealthoughthetotalamoun
tofcovalentbindingwascomparableintheabsenceorpresenceofphosphatethisactivesiteligandpreventedtheinhibitionofenzymaticactivitys
eeninitsabsencethisprotectiveactionofphosphatediminishedwithprogressiveethylationofrnaseindicatingthatthereversibleassociation
ofphosphatewiththeactivesitelysylresiduewasovercomebytheirreversibleprocessofreductiveethylationmodifiedrnasewasanalysedusingc
protondecouplednmrspectroscopypeaksarisingfromthecovalentbindingofenrichedacetaldehydetofreeaminogroupsintheabsenceofphosphate
wereasfollowsnhterminalalphaaminogroupppmbulkethylationatepsilonaminogroupsofnonessentiallysylresiduesppmandtheepsilonaminogro
upoflysineattheactivesiteppminthespectrumofrnaseethylatedinthepresenceofphosphatethepeakatppmwasabsentwhenrnasewasselectivelyp
remethylatedinthepresenceofphosphatetoblockallbuttheactivesitelysylresiduesandthenethylatedinitsabsencethesignalatppmwasgreatl
ydiminishedandthatarisingfromtheactivesitelysylresidueatppmwasenhancedtheseresultsindicatethatphosphatespecificallyprotectedth
eactivesitelysinefromreactionwithacetaldehydeandthatmodificationofthislysinebyacetaldehydeadductformationresultedininhibitiono
fcatalyticactivity
> Reductions in breath ethanol readings in normal male volunteers following mouth rinsing with water at differing temperatures
reductionsinbreathethanolreadingsinnormalmalevolunteersfollowingmouthrinsingwithwateratdifferingtemperaturesbloodethanolconcen
trationsweremeasuredsequentiallyoveraperiodofhoursusingalionaedalcolmeterinhealthymalesubjectsgivenoralethanolgkgbodywtreading
sweretakenbeforeandafterrinsingthemouthwithwateratvaryingtemperaturesmouthrinsingresultedinareductioninthealcolmeterreadingsat
allwatertemperaturestestedthemagnitudeofthereductionwasgreaterafterrinsingwithwateratlowertemperaturesthiseffectoccursbecauser
insingcoolsthemouthanddilutesretainedsalivathisfindingshouldbetakenintoaccountwheneverbreathanalysisisusedtoestimatebloodethan
olconcentrationsinexperimentalsituations
> Does the blockade of opioid receptors influence the development of ethanol dependence?
doestheblockadeofopioidreceptorsinfluencethedevelopmentofethanoldependencewehavetestedwhethertheopioidantagonistsnaloxonemgkgn
altrexonemgkganddiprenorphinemgkgandtheagonistmorphinemgkggivensubcutaneouslyminbeforeethanolfordaysmodifytheethanolwithdrawal
syndromeaudiogenicseizuresfollowingchronicethanolintoxicationinratswefoundthatnaloxonenaltrexoneanddiprenorphinemodifiedtheeth
anolwithdrawalsyndromethesefindingsdonotruleoutthepossibilityofabiochemicallinkbetweentheactionofethanolandopiatesatthelevelof
opioidreceptors
|
Next, the text is read and and broken down into three character "words."
|
Indexing using n-grams words
|
#+++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
#+
#+ Mumps Information Storage and Retrieval Software Library
#+ Copyright (C) 2005, 2007 by Kevin C. O'Kane
#+
#+ Kevin C. O'Kane
#+ okane@cs.uni.edu
#+
#+
#+ This program is free software; you can redistribute it and/or modify
#+ it under the terms of the GNU General Public License as published by
#+ the Free Software Foundation; either version 2 of the License, or
#+ (at your option) any later version.
#+
#+ This program is distributed in the hope that it will be useful,
#+ but WITHOUT ANY WARRANTY; without even the implied warranty of
#+ MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the
#+ GNU General Public License for more details.
#+
#+ You should have received a copy of the GNU General Public License
#+ along with this program; if not, write to the Free Software
#+ Foundation, Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
#+
#++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
# shredder.mps March 28, 2007
open 1:"osu.fasta,old"
set doc=1
for do
. use 1
. read a
. if '$t break
. set ^t(doc)=a
. read a
. for do
.. set word=$zShred(a,3)
.. if word="" break
.. set ^doc(doc,word)=""
. set doc=doc+1
set %=$zzTranspose(^doc,^index)
|
The following is a simple program to retrieve text
based on 3 character n-grams. Note that the ShredQuery()
function produces overlapping n-grams from the
query.
|
Program to Retrieve Text Based on 3 letter Encodings
|
#+++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
#+
#+ Mumps Information Storage and Retrieval Software Library
#+ Copyright (C) 2005 by Kevin C. O'Kane
#+
#+ Kevin C. O'Kane
#+ okane@cs.uni.edu
#+
#+
#+ This program is free software; you can redistribute it and/or modify
#+ it under the terms of the GNU General Public License as published by
#+ the Free Software Foundation; either version 2 of the License, or
#+ (at your option) any later version.
#+
#+ This program is distributed in the hope that it will be useful,
#+ but WITHOUT ANY WARRANTY; without even the implied warranty of
#+ MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the
#+ GNU General Public License for more details.
#+
#+ You should have received a copy of the GNU General Public License
#+ along with this program; if not, write to the Free Software
#+ Foundation, Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
#+
#++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
# shredquery.mps 11/15/05
read "query: ",query
for do
. if $l(query)=0 break
. set word=$$^ShredQuery(query,3)
. if word="" break
. set d=""
. for do
.. set d=$order(^index(word,d))
.. if d="" break
.. if $data(^result(d)) set ^result(d)=^result(d)+1
.. else set ^result(d)=1
set d=""
for do
. set d=$order(^result(d))
. if d="" break
. set ^ans($justify(^result(d),5),d)=""
set sc=""
set %=0
for do
. set sc=$order(^ans(sc),-1)
. if sc="" break
. set d=""
. for do
.. set d=$order(^ans(sc,d))
.. if d="" break
.. write sc," ",d," ",^title(d),!
.. set %=%+1
.. if %>20 halt
|
The above produces output of the form (longer titles truncated):
|
Example Output
|
|
sidhe:/r0/MEDLINE-TMP # shredquery.cgi
query: alcohol
10 100214 > Lithium treatment of depressed and nondepressed alcoholics [see comments]
10 100502 > Alcohol effects on luteinizing hormone releasing hormone-stimulated anterior pituitary and gonadal hormones in women.
10 100656 > Is alcohol consumption related to breast cancer? Results from the Framingham Heart Study.
10 101146 > Hyaluronic acid and type III procollagen peptide in jejunal perfusion fluid as markers of connective tissue turnover.
10 101401 > Prevalence, detection, and treatment of alcoholism in hospitalized patients [see comments]
10 10210 > Alcoholic intemperance, coronary heart disease and mortality in middle-aged Swedish men.
10 102107 > Functional and structural changes in parotid glands of alcoholic cirrhotic patients.
10 103730 > Alcohol consumption and mortality in aging or aged Finnish men [published erratum appears in J Clin Epidemiol 1989;42(7):701]
10 103762 > The role of liquid diet formulation in the postnatal ethanol exposure of rats via mother's milk.
10 103913 > Comparative effectiveness and costs of inpatient and outpatient detoxification of patients with mild-to-moderate alcohol withdrawal ...
10 103926 > The effects of alcoholism on skeletal and cardiac muscle [see comments]
10 10407 > Genetic models of alcohol dependence.
10 10411 > Genetic control of liver alcohol dehydrogenase expression in inbred mice.
10 104287 > The generation of acetonemia/acetonuria following ingestion of a subtoxic dose of isopropyl alcohol.
10 10439 > Increased alcohol intake induced by chronic stimulants: is "orality" involved?
10 10440 > Naloxone attenuation of voluntary alcohol consumption.
10 10441 > Neonatal antidepressant administration suppresses concurrent active (REM) sleep and increases adult alcohol consumption in rats.
10 10444 > Is carbohydrate metabolism genetically related to alcohol drinking?
10 10449 > The antisocial and the nonantisocial male alcoholic--II.
10 10450 > Alcohol and ambience: social and environmental determinants of intake and mood.
10 10454 > Life events in the biography of French male and female alcoholics.
|
The above example generates very large indexing files due to the large number of
fragments extracted for each abstract. To be more effective, the distribution
of the fragments using an algorithm such as the Inverse Document Frequency
method should be used to rank fragments for their likely usefulness in
resolving articles. In practice, as is the case when dealing with
actual natural language text words, fragments whose distributions
are very wide and very narrow can be deleted from the indexing set. (See the
section below on application of IDF to genomic data.)
Overview of Other Methods
Latent Semantic Model
Essentially and term-term co-occurrence method used to augment
queries. An execellent example of a very bad patent
award (1988) - the underlying techniques have been around since the 1960's.
References to Papers on LSI
Wikipedia Entry on LSI
Single Term Based Indexing
Reference: Salton 1983.
Summary:
-
Single term methods view the collection as a set of
individual terms.
-
Methodologies based on this approach
seek to identify which terms are most indicative of content
and to quantify the relative resolving power of each term.
-
Documents are generally viewed as vectors of terms weighted
by the product of a term's frequency of occurrence
in the document and its weight in the collection as a whole.
-
The vector space of a document collection
may be treated as an hyperspace and the effect of
a term on the space density may be taken as indicative of the term's
resolving or discriminating power.
-
Documents may be clustered based on the
similarities of their vector representations.
Hierarchies of clusters may be generated.
-
Queries are treated as weighted vectors and the calculated
similarity between a query vector and document vectors
determines the ranking of a document in the
results presented.
-
Answers to queries are ranked by recall
and precision - measures of the degree of effectiveness of the
methodology to find all documents in a collection
relevant to a query and the degree to which irrelevant
documents are included with relevant documents.
-
Queries may be enhanced by including terms whose
usage patterns are similar to words in the original
query.
Phrase Based Indexing
-
These are based on multiple words taken as phrases
that are likely to convey greater context so as
to improve precision.
-
Methods can involve thesauruses which group multiple
words together into concept classes.
-
Methods can seek the identification of key phrases in a document.
or construction of phrases from document context.
N-Gram Based Indexing
N-grams
Example Code
The following link is to a collection of Mumps code that
performs the operations listed above:
http://www.cs.uni.edu/~okane/source/ISR/ISR115Code.tgz .
Visualization
An increasingly important aspect of IS&R is the ability to
present the results to the user in a meaningful manner and interact
with the user to refine his or her requests. In the early
Salton experiments, queries and results were in text format
only and generally entered, processed, and returned in batches.
Since the widespread availability of the Internet as well
and the general availability of graphical user interfaces,
newer methods of rendering results can be explored.
The following links give examples of several of these:
-
Modern Information Retrieval
Chapter 10: User Interfaces and Visualization - by Marti Hearst
-
Visualizing the Non-Visual: spatial Analysis and Interaction with Information from Text Documents
J.A. Wise, J.J. Thomas, K. Pennock,
D. Lantrip, M. Pottier, A. Schur, and V. Crow (PDF file)
Visualizing the Non-Visual: Spatial Analysis and Interaction with Information from Text Documents
J.A. Wise, J.J. Thomas, K. Pennock,
D. Lantrip, M. Pottier, A. Schur, and V. Crow (PowerPoint File)
-
Internet browsing and searching: User evaluations of category map and Concept Space
(PDF)
-
Exploring the World Wide Web with Self-Organizing Map
-
HIBROWSE interfaces
-
Scatter/Gather clustering
Text Searching
Open Directory
Open Directory Project
Text Searching in Genomic Data Bases
-
Entrez/PubMed
-
SRS
Applications to Genomic Data Bases
In the past 15 years, genomics data bases have grown rapidly. These include
both text and genetic information and the need to search them is central
to research in areas such as genetic, drug discovery and medicine, to name a
few.
Genetic data bases include both natural language text as well as sequences of
genetic information. Data bases containing genetic data bases are primarily
divided into two types: those containing protein information
and those containing DNA sequences. DNA sequence data bases
usually consist of natural language text together with DNA
sequence information over the nucleotide alphabet of bases {ACGT}. These letters stand
for:
- Adenine
- Cytosine
- Guanine
- Thymine
DNA itself is a double stranded helix with the nucleotides constituting
the links across the strands (see: here)
Protein sequence data bases are over the alphabet
{ACDEFGHIKLMNPQRSTVWY} of amino acids.
Sections of DNA are codes that are used
to construct proteins.
The order of the amino acids in a protein determine its shape, chemical activity and function.
DNA substrings become proteins through a process of
translation and transcription. The DNA is divided into three letter codons
which select
amino acids for incorporation into the protein being built.
Over evolutionary time, DNA and resulting protein structures mutate.
Thus, when searching either a protein or nucleotide data base,
exact matches are not always the case. However, it has been observed
that certain mutations are favored in nature while others
are not. Generally speaking, this is because some mutations
do not effect the functionality of the resulting proteins
while other render the protein unusable.
Many searching algorithms take evolutionary mutation into account
through the use of substitution matrices. These give a score
as to the probability of a given substitution of one amino acid
for another based on observation in nature. The
BLAST substitution
matrices are typical. Different matrices are used to
account for the amount of presumed evolutionary distance.
GenBank
"GenBank is a component of a tri-partite, international collaboration of
sequence databases in the U.S., Europe, and Japan. The collaborating
database in Europe is the European Molecular Biology Laboratory (EMBL)
at Hinxton Hall, UK, and in Japan, the DNA Database of Japan (DDBJ) in
Mishima, Japan. Patent sequences are incorporated through arrangements
with the U.S. Patent and Trademark Office, and via the collaborating
international databases from other international patent offices.
The database is converted to various output formats including the Flat File
and Abstract Syntax Notation 1 (ASN.1) versions. The ASN.1 form of the data is
included in www-Entrez and network-Entrez and is also available, as is the
flat file, by anonymous FTP to 'ftp.ncbi.nih.gov'." (ftp://ftp.ncbi.nih.gov/genbank/README.genbank)
- Main GenBank FTP Site
- General Release Notes
- Feature Table Definitions
- Annotated Sample Record
EMBL/EBI (European Molecular Biology Laboratory/European Bioinformatics Institute
"The European Bioinformatics Institute (EBI) is a non-profit academic organisation that forms part of the European Molecular Biology Laboratory (EMBL).
The EBI is a centre for research and services in bioinformatics. The Institute manages databases of biological data including nucleic acid, protein sequences and macromolecular structures.
The mission of the EBI is to ensure that the growing body of information from molecular biology and genome
research is placed in the public domain and is accessible freely to all facets of the scientific community in
ways that promote scientific progress."
(from: http://www.ebi.ac.uk/Information/ )
- EBI Home Page
- EBI FTP Server
Alignment Algorithms
In bioinformatics, a researcher identifies a protein or nucleotide sequence
and wants to locate similar sequences in the data base. Some
of the earliest methods used involve sequence alignment.
Most direct sequence alignment techniques are very compute
intensive and impractical to be applied with very
large data bases. However, they represent the "gold standard"
for sequence matching.
The following is an historical overview of alignment algorithms
and their evolution.
Mumps Smith-Waterman Example
FASTA
BLAST (Basic Local Alignment Sequencing Tool)
- Blast HTML home page
- Blast FTP home page
- Blast Data Bases
- Substitution Matrices
- Blast Executables
- Blast Algorithm
Case Study: Indexing the "nt" Data Base
This section explores the hypothesis that it is possible to identify genomic sequence fragments in
large data bases whose indexing characteristics are comparable to that of a weighted vocabulary of
natural language words. The Inverse Document Frequency (IDF) is a simple but widely used natural language
word weighting factor that measures the relative importance of words in a collection based on
word distribution. A high IDF weight usually indicates an important content descriptor.
An experiment was conducted to calculate the relative IDF weights of all segmented non-overlapping
fixed length n-grams of length eleven in the NCBI "nt" and other data bases. The
resulting n-grams were ranked by weight; the effect on sequence retrieval calculated in
randomized tests; and the results compared with BLAST and MegaBlast for accuracy and speed. Also
discussed are several anomalous specific weight distributions indicative of differences
in evolutionary vocabulary.
BLAST and other similar systems pre-index each data base sequence by short code letter words of, by
default, three letters for data bases consisting of strings over the larger amino acid alphabet and
eleven letters for data bases consisting of strings over the four character nucleotide alphabet. Queries
are decomposed into similar short code words. In BLAST, the data base index is sequentially scanned and
those stored sequences having code words in common with the query are processed further to extend the initial
code word matches. Substitution matrices are often employed to accommodate mutations due to evolutionary
distance and statistical analyses predict if an alignment is by chance, relative to the size of the data base.
Indexing and retrieving natural language text presents similar problems. Both areas deal with very large
collections of text material, large vocabularies and a need to locate information based on imprecise and
incomplete descriptions of the data. With natural language text, the problem is to locate those documents
that are most similar to a text query. This, in part, can be accomplished by techniques that identify those
terms in a document collection that are likely to be good indicators of content. Documents are converted to
weighted vectors of these terms so as to position each document in an n-dimensional hyperspace
where "n" is the number of terms. Queries are likewise converted to vectors of terms to denote a point in the hyperspace and documents ranked as possible answers to the query by one of several well known formulas to measure the distance of a document from a query. Natural language systems also employ extensive inverted file structures where content is addressed by multiple weighted descriptors.
During World War II, n-grams, fixed length consecutive series of "n" characters, were developed by
cryptographers to break substitution ciphers. Applying n-grams to indexing, the text, stripped of
non-alphabetic characters, is treated as a continuous stream of data that is segmented into
non-overlapping fixed length words. These words can then form the basis of the indexing vocabulary.
The purpose of this experiment was to determine if it were possible to computationally identify genomic sequence
fragments in large data bases whose indexing characteristics are similar to that of a weighted vocabulary of natural
language words. The experiments employed an n-gram based information retrieval system utilizing an inverse document frequency
(IDF) term weight and an incidence scoring methodology. The results were compared with BLAST and MegaBlast to
determine if this approach produced results of comparable recall when retrieving sequences from the data base
based on mutated and incomplete queries.
This experimental model incorporates no evolutionary assumptions and is based entirely on a computational analysis
the contents of the data base. That is, this approach does not, by default, use any substitution matrices or
sequence translations. The software does, however, allow the inclusion of a file of aliases, effectively substitutions
and translations are always a possible extra step. The distribution package includes a module that can compute
possible aliases based on term-term correlations or on well known empirically based amino acid substitutions.
Experiment Design
For our primary experiments, sequences from the very large NCBI "nt" non-redundant nucleotide data base were
used. The "nt" data base (ftp://ftp.ncbi.nih.gov/blast/db/FASTA) was approximately 12 billion bytes in length at
the time of the experiment and consisted 2,584,440 sequences in FASTA format. Other experiments using the
nucleotide primate, est, plant, bacteria, viral, rodent and other collections in GenBank were also performed as noted
below.
|
Example Entries from "nt" (FASTA Format)
|
>gi|2695846|emb|Y13255.1|ABY13255 Acipenser baeri mRNA for immunoglobulin heavy chain, clone ScH 3.3
TGGTTACAACACTTTCTTCTTTCAATAACCACAATACTGCAGTACAATGGGGATTTTAACAGCTCTCTGTATAATAATGA
CAGCTCTATCAAGTGTCCGGTCTGATGTAGTGTTGACTGAGTCCGGACCAGCAGTTATAAAGCCTGGAGAGTCCCATAAA
CTGTCCTGTAAAGCCTCTGGATTCACATTCAGCAGCGCCTACATGAGCTGGGTTCGACAAGCTCCTGGAAAGGGTCTGGA
ATGGGTGGCTTATATTTACTCAGGTGGTAGTAGTACATACTATGCCCAGTCTGTCCAGGGAAGATTCGCCATCTCCAGAG
ACGATTCCAACAGCATGCTGTATTTACAAATGAACAGCCTGAAGACTGAAGACACTGCCGTGTATTACTGTGCTCGGGGC
GGGCTGGGGTGGTCCCTTGACTACTGGGGGAAAGGCACAATGATCACCGTAACTTCTGCTACGCCATCACCACCGACAGT
GTTTCCGCTTATGGAGTCATGTTGTTTGAGCGATATCTCGGGTCCTGTTGCTACGGGCTGCTTAGCAACCGGATTCTGCC
TACCCCCGCGACCTTCTCGTGGACTGATCAATCTGGAAAAGCTTTT
>gi|2695850|emb|Y13260.1|ABY13260 Acipenser baeri mRNA for immunoglobulin heavy chain, clone ScH 16.1
TCTGCTGGTTACAACACTTTCTTCTTTCAATAACCACAATACTGCAGTACAATGGGGATTTTAACAGCTCTCTGTATAAT
AATGACAGCTCTATCAAGTGTCCGGTCTGATGTAGTGTTGACTGAGTCCGGACCAGCGGTTGTAAAGCCTGGAGAGTCCC
ATAAACTGTCCTGTAAAGCCGCTGGATTCACATTCAGCAGCTATTGGATGGGCTGGGTTCGACAAACTCCGGGAAAGGGT
CTGGAATGGGTGTCTATTATAAGTGCTGGTGGTAGTACATACTATGCCCCGTCTGTTGAGGGACGATTCACCATCTCCAG
AGACAATTCCAACAGCATGCTGTATTTACAAATGAACAGCCTGAAGACTGAAGACACTGCCATGTATTACTGTGCCCGCA
AACCGGAAACGGGTAGCTACGGGAACATATCTTTTGAACACTGGGGGAAAGGAACAATGATCACCGTGACTTCGGCTACG
CCATCACCACCGACAGTGTTTCCGCTTATGCAGGCATGTTGTTCGGTCGATGTCACGGGTCCTAGCGCTACGGGCTGCTT
AGCAACCGAATTC
>gi|2695852|emb|Y13263.1|ABY13263 Acipenser baeri mRNA for immunoglobulin heavy chain, clone ScH 112
CAAGAACCACAATACTGCAGTACAATGGGGATTTTAACAGCTCTCTGTATAATAATGACAGCTCTATCAAGTGTCCGGTC
TGATGTAGTGTTGACTGAGTCCGGACCAGCAGTTATAAAGCCTGGAGAGTCCCATAAACTGTCCTGTAAAGCCTCTGGAT
TCACATTCAGCAGCAACAACATGGGCTGGGTTCGACAAGCTCCTGGAAAGGGTCTGGAATGGGTGTCTACTATAAGCTAT
AGTGTAAATGCATACTATGCCCAGTCTGTCCAGGGAAGATTCACCATCTCCAGAGACGATTCCAACAGCATGCTGTATTT
ACAAATGAACAGCCTGAAGACTGAAGACTCTGCCGTGTATTACTGTGCTCGAGAGTCTAACTTCAACCGCTTTGACTACT
GGGGATCCGGGACTATGGTGACCGTAACAAATGCTACGCCATCACCACCGACAGTGTTTCCGCTTATGCAGGCATGTTGT
TCGGTCGATGTCACGGGTCCTAGCGCTACGGGCTGCTTAGCAACCGAATTC
>gi|2695854|emb|Y13264.1|ABY13264 Acipenser baeri mRNA for immunoglobulin heavy chain, clone ScH 113
TTTCTTCTTTCAATAACCACAATACTGCAGTACAATGGGGATTTTAACAGCTCTCTGTATAATAATGACAGCTCTATCAA
GTGTCCAGTCTGATGTAGTGTTGACTGAGTCCGGAACAGCAGTTATAAAGCCTGGAGAGTCCCATAAACTGTCCTGTAAA
GCCTCTGGATTCACATTCAGCAGCTACTGGATGGGCTGGGTTCGACAAGCTCCTGGAAAGGGTCTGGAATGGGTGTCTAC
TATAAGCAGTGGTGGTAGTGCGACATACTATGCCCCGTCTGTCCAGGGAAGATTCACCATCTCCAGAGACGATTCCAACA
GCCTGCTGTCTTTACAAATGAACAGCCTGAAGACTGAAGACACTGCCGTCTATTACTGTGCTCGAAACTTACGGGGGTAC
GAGGCTTTCGACCTCTGGGGTAAAGGGACCATGGTCACCGTAACTTCTGCTACGCCATCACCACCGACAGTGTTTCCGCT
TATGCAGGCATGTTGTTCGGTCGATGTCACGGGTCCTAGCGCTACGGGCTGCTTAGCAACCGAATTC
|
The overall frequencies of occurrence of all possible non-overlapped 11 character words in each sequence in the data base were determined along with the number of sequences in which each unique word was found. A total of 4,194,299 unique words were identified, slightly less than the theoretical maximum of 4,194,304. The word size of 11 was initially selected as this is the default word size used in BLAST for nucleotide searches. The programs however, will accommodate other word lengths and the default size for proteins is three.
Each sequence in the "nt" data base was read and decomposed into all possible words of length 11. Procedurally, given the vast amount of words thus produced, multiple (about 110 in the case of "nt") intermediate files of about 440 million bytes each were produced. Each file was ordered alphabetically by word and listing, for each word, a four byte relative reference number of the original sequence containing the word. Another table was also produced that translated each relative reference number to an eight byte true offset into the original data base. The multiple intermediate files were subsequently merged and three files produced: (1) a large (40 GB) ordered word-sequence
master table giving, for each word, a list of the sequence references of those sequences in which the word occurs; (2) a file containing the IDF weights for each word; and (3) a file giving for each word the eight byte offset of the word's entry in the master table.
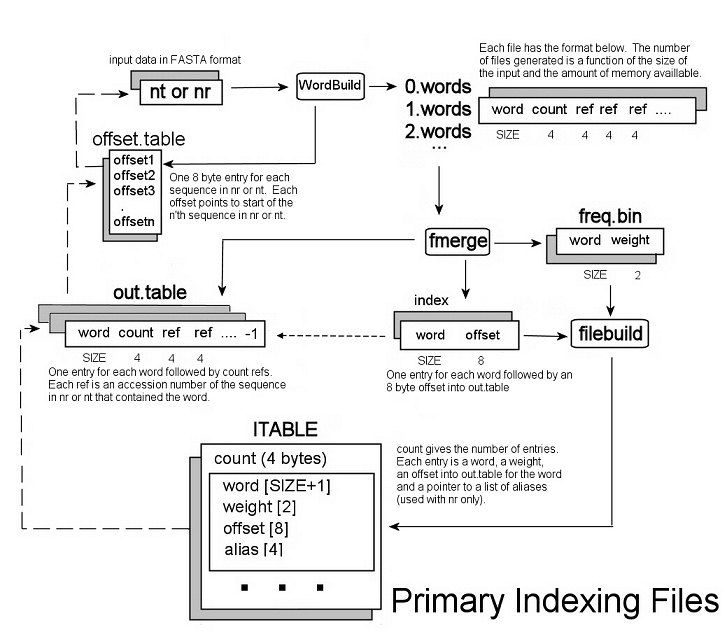
Flowchart 1
Source code copies of this code are available at:
http://www.cs.uni.edu/~okane/source/
in the file
named idf.src-1.06.tar.gz (note: version number will change with time).
The IDF weights (freq.bin) Wi for each word i were calculated by:
Wi= (int) 10 * Log10 ( N / DocFreqi ) (1)
where N is the total number of sequences, and DocFreq is the total number of sequences in which each word occurred. This weight yields higher values for words whose distribution is more concentrated and lower values for words whose use is more widespread. Thus, words of broad context are weighted lower than words of narrow context.
For information retrieval, each query sequence was read and decomposed into overlapping 11 character words which were converted to a numeric equivalent for indexing purposes. Entries in a master scoring vector corresponding to data base sequences were incremented by the weight of the word if the word occurred in the sequence and if the weight of the word lay within a specified range. When all words had been processed, entries in the master sequence vector were normalized according to the length of the underlying sequences and to the length of the query. Finally, the master sequence vector was sorted by total weight and the top scoring entries were either displayed with IDF based weights, or scored and ranked by a built-in Smith-Waterman alignment procedure.
Results
All tests were conducted on a dual processor Intel Xeon 2.25 mHz system with 4 GB of memory and 5,500 rpm disk drives operating under Mandrake Linux 9.2. Both software systems benefited from the large memory to buffer I/O requests but BLAST, due to the more compact size of its indexing files (about 3 GB vs. 40 GB), was able to load a very substantially larger percentage of its data base into memory which improved its performance in serial trials subsequent to the first.
Figure 1 shows a graph of aggregate word frequency by weight. The height of each bar reflects the total number of instances of all words of a given weight in the data base. The bulk of the words, as is also the case with natural language text3,7, reside in the middle range.
Figure 1
Initially, five hundred test queries were randomly generated from the "nt" data base by (1) randomly selecting sequences whose length was between 200 and 800 letters; (2) from each of these, extracting a random contiguous subsequence between 200 and 400 letters; and (3) randomly mutating an average of 1 letter out of 12. While this appears to be a small level of mutation, it is significant for both BLAST and IDF where the basic indexing word size is, by default, 11. A "worst case" of mutation for either approach would be a sequence in which each word were mutated. In our mutation procedure, each letter of a sequence had a 1 in 12 chance of being mutated.
The test queries were processed and scored by the indexing program with IDF weighting enabled and disabled and also by BLAST. The output of each consisted of 500 sequence title lines ordered by score. The results are summarized in Table 1 and Figures 2 and 3. In Figures 2 and 3, larger bars further to the left indicate better performance (ideally, a single large bar at position 1). The Average Time includes post processing of the results by a Perl program. The Average Rank and Median Rank refer to the average and median positions, respectively, in the output of the sequence from which a query was originally derived. A lower number indicates better performance. The bar at position 60 indicates all ranks 60 and above as well as sequences not found.
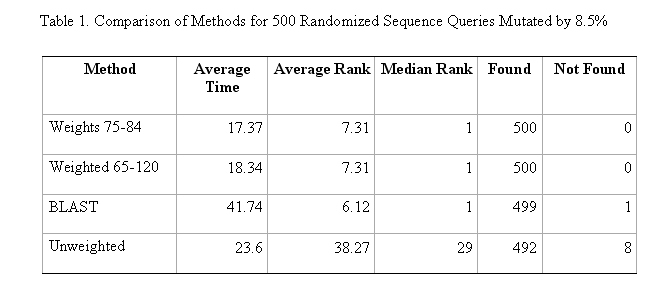
Table 1
Figure 2
Figure 3
When running in unweighted mode, all words in a query were weighted equally and sequences containing those words were scored exclusively on the unweighted cumulative count of the words in common with the query vector. When running in weighted mode, query words were used for indexing if they fell within the range of weights being tested and data base sequences were scored on the sum of the weights of the terms in common with the query vector and normalized for length.
Figure 3 shows results obtained using the 500 random sequences using indexing only and no weights. The graph in Figure 2 shows significantly better results for the same query sequences with weighted indexing enabled (see also Table 1).
Subsequently, multiple ranges of weights were tested with the same random sequences. In these tests, only words within certain weight ranges were used. The primary indicators of success were the Average Rank and the number of sequences found and not found. From these results, optimal performance was obtained using weights in the general range of 65 to 120. The range 75 to 84 also yielded similar information retrieval performance with slightly better timing.
Table 2 shows the results of a series of trials at various levels of mutation and query sequence length. The numbers indicate the percentage of randomly generated and mutated queries of various lengths found. The IDF method is comparable to BLAST at mutations of 20% or less. In all cases, the IDF method was more than twice as fast.
Table 2
On larger query sequences (5,000 to 6,000 letters), the IDF weighted method performed slightly better than BLAST. On 25 long sequences randomly generated as noted above, the IDF method correctly ranked the original sequence first 24 times, and once at rank 3. BLAST, on the other hand, ranked the original sequence first 21 times while the remaining 4 were ranked 2, 2, 3 and 4. Average time per query for the IDF method was 47.4 seconds and the average time for BLAST was 122.8 seconds.
Word sizes other than eleven were tested but with mixed results. Using a word longer than eleven greatly increases the number of words and intermediate file sizes while a smaller value results in too few words relative the number of sequences to provide full resolution.
A set of random queries was also run against MegaBlast. MegaBlast is a widely used fast search procedure that employs a greedy algorithm and is dependent upon larger word sizes (28 by default). The results of these trials were that the IDF method was able to successfully identify all candidates while MegaBlast failed to identify any candidates. MegaBlast is primarily useful in cases where the candidate sequences are a good match for a target database sequence.
Figure 4 is a graph of the number of distinct words at each weight in the "nt" data base. The twin peaks were unexpected. The two distinct peaks suggest the possible presence of two .vocabularies. with overlapping bell curves. To test this, we separately indexed the nucleotide data in the NCBI GenBank collections for primates (gbpri*), rodents (gbrod*), bacteria (gbbct*), plants (gbpln*), vertebrates (gbvrt*), invertebrates (gbinv*), patented sequences (gbpat*), viruses (gbvir*), yeast (yeast_gb.fasta) and phages (gbphg*) and constructed similar graphs. The virus, yeast, and phage data bases were too small to give meaningful results and the patents data base covered many species. The other databases, however yielded the graphs shown in Figure 5 which, for legibility, omits vertebrates and invertebrates (see below). In this figure, the composite NT data base graph is seen with the twin peaks as noted from Figure 4. Also seen are the primate and rodent graphs which have similar but more pronounced curves. The curves for bacteria and plants display single peaks. The invertebrate graph is roughly similar to the bacteria and plant graphs and the vertebrate curve is roughly similar to primates and rodents although both these data sets are small and the curves are not well defined.
Figure 4
Figure 5
The origin and significance of the twin peaks is not fully understood. It was initially hypothesized that it may be due to mitochondrial DNA in the samples. To determine if this were the case, the primate data base was stripped of all sequences whose text description used the term .mitochon*.. This removed 19,647 sequences from the full data bases of 334,537 sequences. The data base was then re-indexed and the curves examined. The curves were unchanged except for a very slight displacement due to a smaller data base (see below). In another experiment, words in a band at each peak in the primate data base were extracted, concatenated, and entered as (very large) queries to the "nt" data base. The resulting sequences retrieved showed some clustering with mouse and primate sequences at words from band 67 to 71 and bacteria more common at band 79 to 83.
The "nt", primate and rodent graphs, while otherwise similar, are displaced from one another as are the plant and bacteria graphs. These displacements appear mainly to be due to differences in the sizes of the data bases and the consequent effect on the calculation of the logarithmic weights. The NT data base at 12 GB is by far the largest, the primate and rodent data set are 4.2 GB and 2.3GB respectively, while the plant and bacteria databases are somewhat similar at 1.4 GB and 0.97 GB, respectively.
Conclusions
The results indicate that it is possible to identify a vocabulary of useful fragment sequences using an n-gram based inverse document frequency weight. Further, an information retrieval system based on this method and incidence scoring is effective in retrieving genomic sequences and is generally better than twice as fast as BLAST and of comparable accuracy when mutations do not exceed 20%. The results also indicate that this procedure works where other speedup methods such as MegaBlast do not.
Significantly, these results imply that genomic sequences are susceptible to procedures used in natural language indexing and information retrieval. Thus, since IDF or similar weight based systems are often at the root of many natural language information retrieval systems, other more computationally intense text natural language indexing, information retrieval and visualization techniques such as term discrimination, hierarchical sequence clustering, synonym recognition, and vocabulary clustering to name but a few, may also be effective and useful.
Miscellaneous Links
-
Survey of techniques - Developments in Automatic Information Retrieval by
G. Salton
-
Origins of language
-
Irregular English Verbs
-
Open Directory - Information Retrieval
Some related lecture slides from UC Berkeley (SIMS 202
Information Organization and Retrieval
Instructors: Marti Hearst & Ray Larson)
-
Introduction to Content Analysis
-
Introduction to Content Analysis Continued
-
Term Weighting and Ranking Algorithms
-
Ranked Retrieval Systems